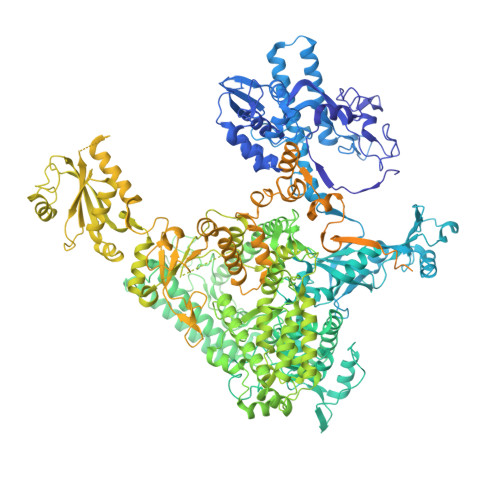
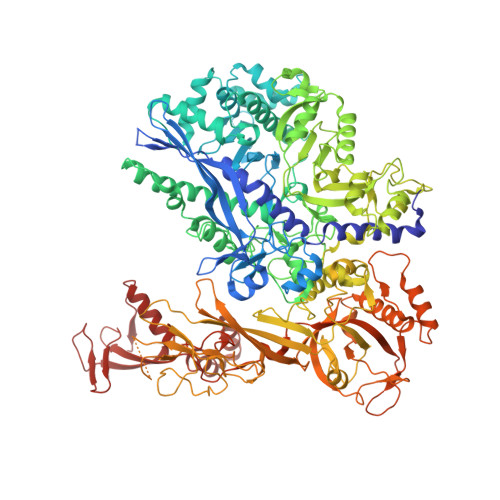
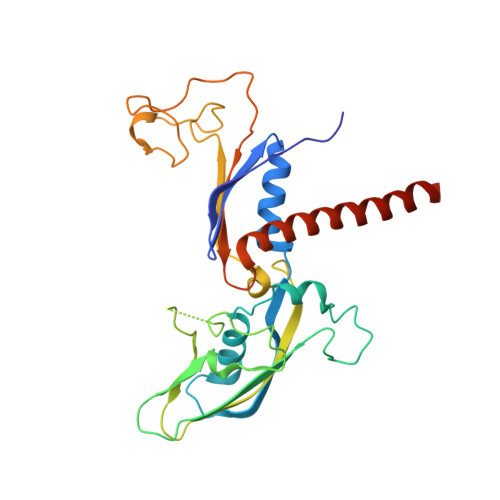
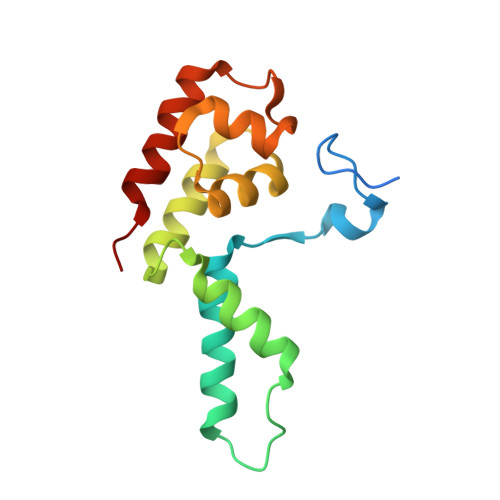
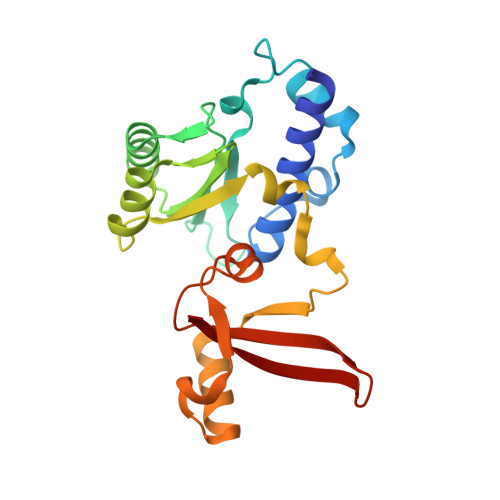
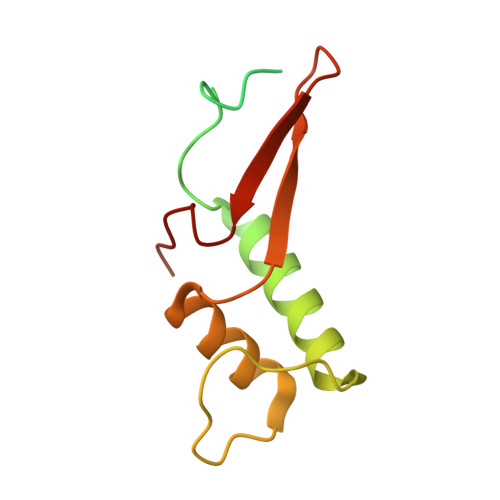
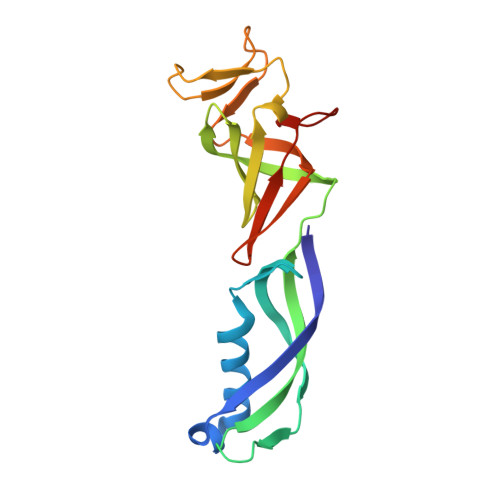
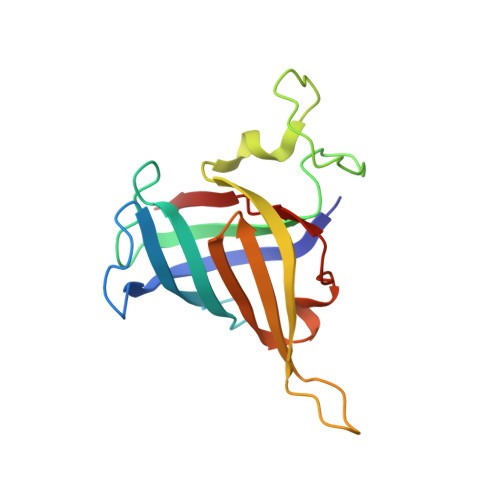
Structural basis of human transcription-DNA repair coupling.
Kokic, G., Wagner, F.R., Chernev, A., Urlaub, H., Cramer, P.(2021) Nature 598: 368-372
- PubMed: 34526721
- DOI: https://doi.org/10.1038/s41586-021-03906-4
- Primary Citation of Related Structures:
7OO3, 7OOB, 7OOP, 7OPC, 7OPD - PubMed Abstract:
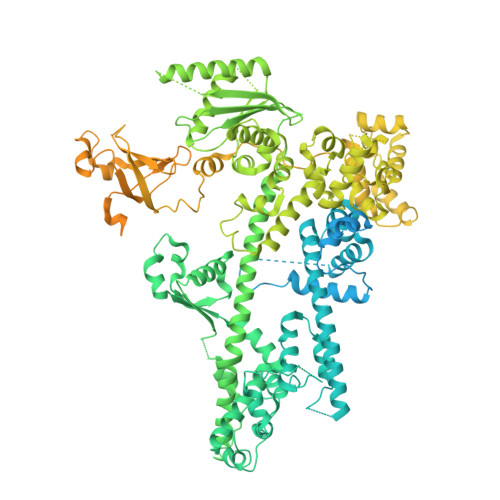
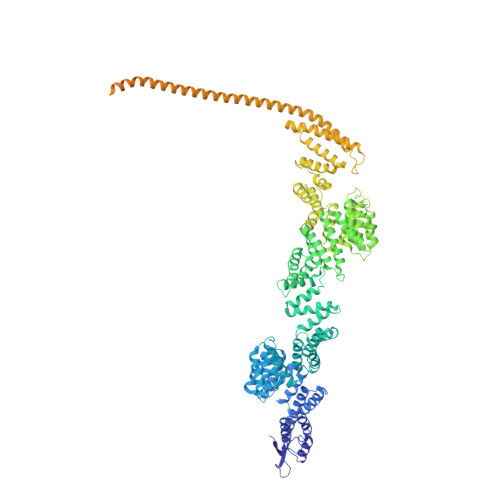
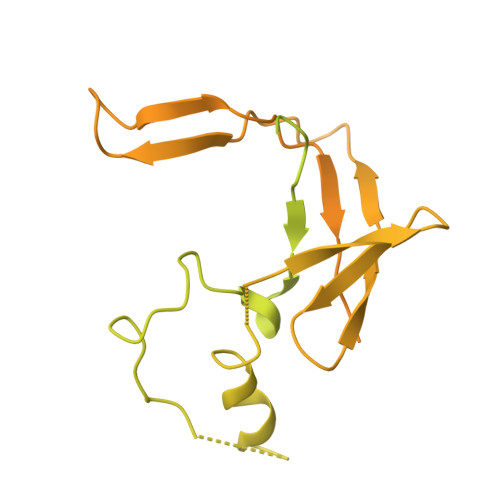
Transcription-coupled DNA repair removes bulky DNA lesions from the genome 1,2 and protects cells against ultraviolet (UV) irradiation 3 . Transcription-coupled DNA repair begins when RNA polymerase II (Pol II) stalls at a DNA lesion and recruits the Cockayne syndrome protein CSB, the E3 ubiquitin ligase, CRL4 CSA and UV-stimulated scaffold protein A (UVSSA) 3 . Here we provide five high-resolution structures of Pol II transcription complexes containing human transcription-coupled DNA repair factors and the elongation factors PAF1 complex (PAF) and SPT6. Together with biochemical and published 3,4 data, the structures provide a model for transcription-repair coupling. Stalling of Pol II at a DNA lesion triggers replacement of the elongation factor DSIF by CSB, which binds to PAF and moves upstream DNA to SPT6. The resulting elongation complex, EC TCR , uses the CSA-stimulated translocase activity of CSB to pull on upstream DNA and push Pol II forward. If the lesion cannot be bypassed, CRL4 CSA spans over the Pol II clamp and ubiquitylates the RPB1 residue K1268, enabling recruitment of TFIIH to UVSSA and DNA repair. Conformational changes in CRL4 CSA lead to ubiquitylation of CSB and to release of transcription-coupled DNA repair factors before transcription may continue over repaired DNA.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany.
Organizational Affiliation: