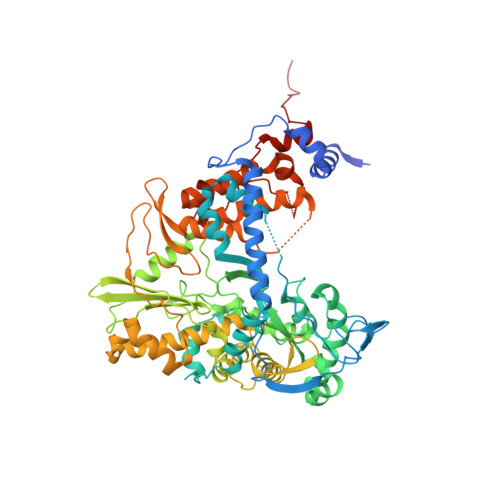
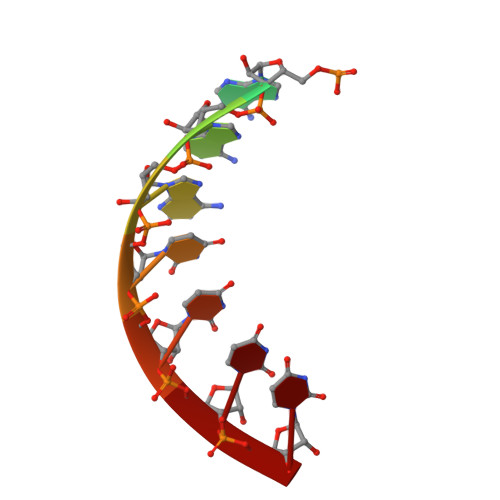
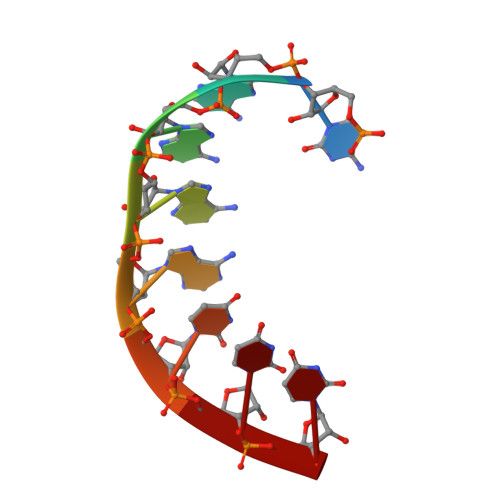
Snapshots of a Non-Canonical RdRP in Action.
Ferrero, D.S., Falqui, M., Verdaguer, N.(2021) Viruses 13
- PubMed: 34203380
- DOI: https://doi.org/10.3390/v13071260
- Primary Citation of Related Structures:
7OM2, 7OM6, 7OM7, 7OM9, 7OMA - PubMed Abstract:
RNA viruses typically encode their own RNA-dependent RNA polymerase (RdRP) to ensure genome replication and transcription. The closed "right hand" architecture of RdRPs encircles seven conserved structural motifs (A to G) that regulate the polymerization activity. The four palm motifs, arranged in the sequential order A to D, are common to all known template dependent polynucleotide polymerases, with motifs A and C containing the catalytic aspartic acid residues. Exceptions to this design have been reported in members of the Permutotetraviridae and Birnaviridae families of positive single stranded (+ss) and double-stranded (ds) RNA viruses, respectively. In these enzymes, motif C is located upstream of motif A, displaying a permuted C-A-B-D connectivity. Here we study the details of the replication elongation process in the non-canonical RdRP of the Thosea asigna virus (TaV), an insect virus from the Permutatetraviridae family. We report the X-ray structures of three replicative complexes of the TaV polymerase obtained with an RNA template-primer in the absence and in the presence of incoming rNTPs. The structures captured different replication events and allowed to define the critical interactions involved in: (i) the positioning of the acceptor base of the template strand, (ii) the positioning of the 3'-OH group of the primer nucleotide during RNA replication and (iii) the recognition and positioning of the incoming nucleotide. Structural comparisons unveiled a closure of the active site on the RNA template-primer binding, before rNTP entry. This conformational rearrangement that also includes the repositioning of the motif A aspartate for the catalytic reaction to take place is maintained on rNTP and metal ion binding and after nucleotide incorporation, before translocation.
- Institut de Biología Molecular de Barcelona, CSIC, Parc Científic de Barcelona, Baldiri Reixac 15, 08028 Barcelona, Spain.
Organizational Affiliation: