Structures and implications of TBP-nucleosome complexes.
Wang, H., Xiong, L., Cramer, P.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34301908
- DOI: https://doi.org/10.1073/pnas.2108859118
- Primary Citation of Related Structures:
7OH9, 7OHA, 7OHB, 7OHC - PubMed Abstract:
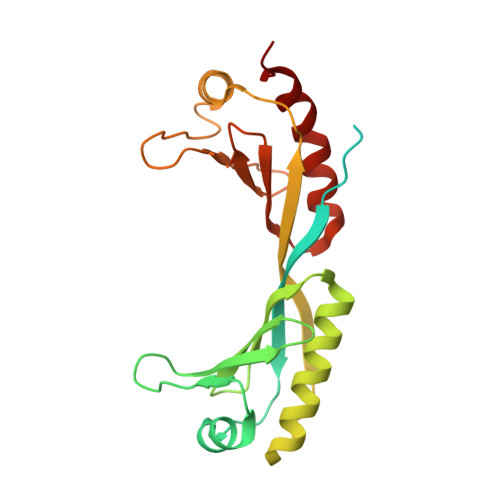
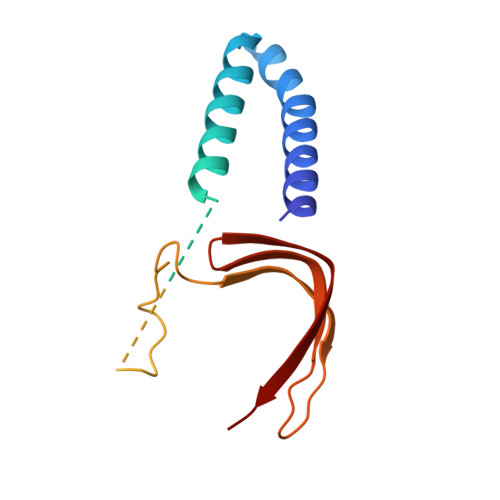
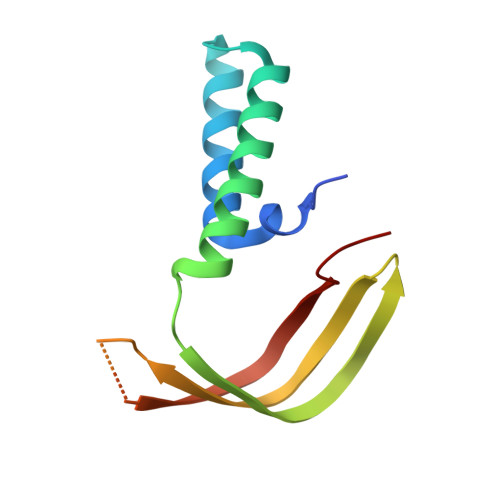
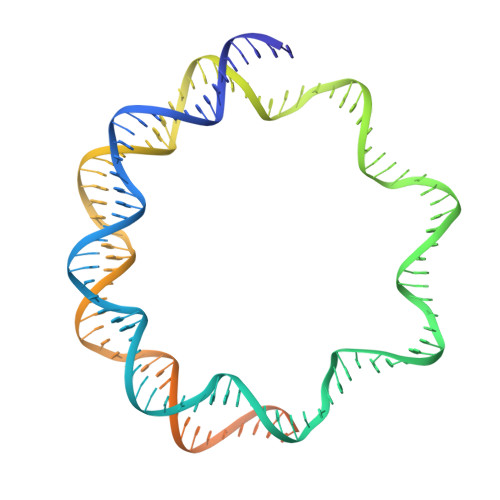
The TATA box-binding protein (TBP) is highly conserved throughout eukaryotes and plays a central role in the assembly of the transcription preinitiation complex (PIC) at gene promoters. TBP binds and bends DNA, and directs adjacent binding of the transcription factors TFIIA and TFIIB for PIC assembly. Here, we show that yeast TBP can bind to a nucleosome containing the Widom-601 sequence and that TBP-nucleosome binding is stabilized by TFIIA. We determine three cryo-electron microscopy (cryo-EM) structures of TBP-nucleosome complexes, two of them containing also TFIIA. TBP can bind to superhelical location (SHL) -6, which contains a TATA-like sequence, but also to SHL +2, which is GC-rich. Whereas binding to SHL -6 can occur in the absence of TFIIA, binding to SHL +2 is only observed in the presence of TFIIA and goes along with detachment of upstream terminal DNA from the histone octamer. TBP-nucleosome complexes are sterically incompatible with PIC assembly, explaining why a promoter nucleosome generally impairs transcription and must be moved before initiation can occur.
- Department of Molecular Biology, Max Planck Institute for Biophysical Chemistry, 37077 Göttingen, Germany.
Organizational Affiliation: