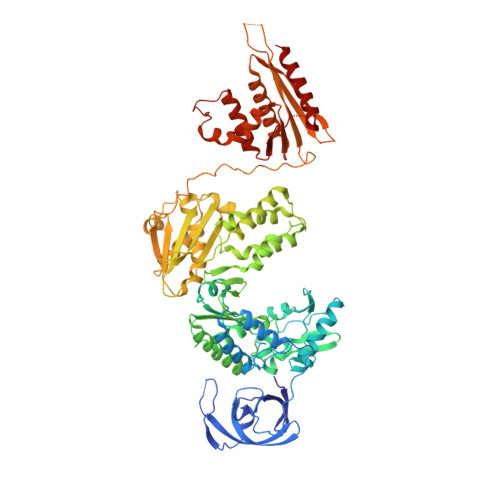
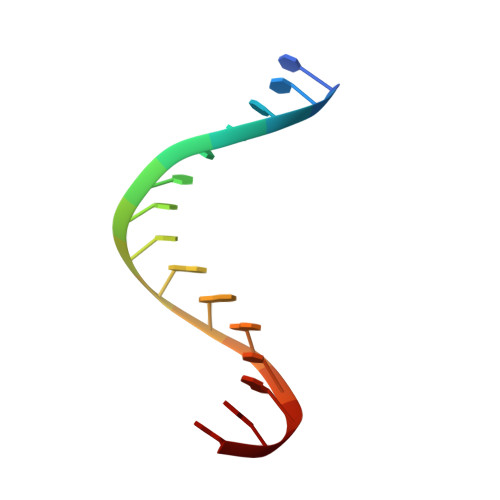
Structures of Substrate Complexes of Foamy Viral Protease-Reverse Transcriptase.
Nowacka, M., Nowak, E., Czarnocki-Cieciura, M., Jackiewicz, J., Skowronek, K., Szczepanowski, R.H., Wohrl, B.M., Nowotny, M.(2021) J Virol 95: e0084821-e0084821
- PubMed: 34232702
- DOI: https://doi.org/10.1128/JVI.00848-21
- Primary Citation of Related Structures:
7O0G, 7O0H, 7O24 - PubMed Abstract:
Reverse transcriptases (RTs) use their DNA polymerase and RNase H activities to catalyze the conversion of single-stranded RNA to double-stranded DNA (dsDNA), a crucial process for the replication of retroviruses. Foamy viruses (FVs) possess a unique RT, which is a fusion with the protease (PR) domain. The mechanism of substrate binding by this enzyme has been unknown. Here, we report a crystal structure of monomeric full-length marmoset FV (MFV) PR-RT in complex with an RNA/DNA hybrid substrate. We also describe a structure of MFV PR-RT with an RNase H deletion in complex with a dsDNA substrate in which the enzyme forms an asymmetric homodimer. Cryo-electron microscopy reconstruction of the full-length MFV PR-RT-dsDNA complex confirmed the dimeric architecture. These findings represent the first structural description of nucleic acid binding by a foamy viral RT and demonstrate its ability to change its oligomeric state depending on the type of bound nucleic acid. IMPORTANCE Reverse transcriptases (RTs) are intriguing enzymes converting single-stranded RNA to dsDNA. Their activity is essential for retroviruses, which are divided into two subfamilies differing significantly in their life cycles: Orthoretrovirinae and Spumaretrovirinae . The latter family is much more ancient and comprises five genera. A unique feature of foamy viral RTs is that they contain N-terminal protease (PR) domains, which are not present in orthoretroviral enzymes. So far, no structural information for full-length foamy viral PR-RT interacting with nucleic substrates has been reported. Here, we present crystal and cryo-electron microscopy structures of marmoset foamy virus (MFV) PR-RT. These structures revealed the mode of binding of RNA/DNA and dsDNA substrates. Moreover, unexpectedly, the structures and biochemical data showed that foamy viral PR-RT can adopt both a monomeric configuration, which is observed in our structures in the presence of an RNA/DNA hybrid, and an asymmetric dimer arrangement, which we observed in the presence of dsDNA.
- Laboratory of Protein Structure, International Institute of Molecular and Cell Biologygrid.419362.b, Warsaw, Poland.
Organizational Affiliation: