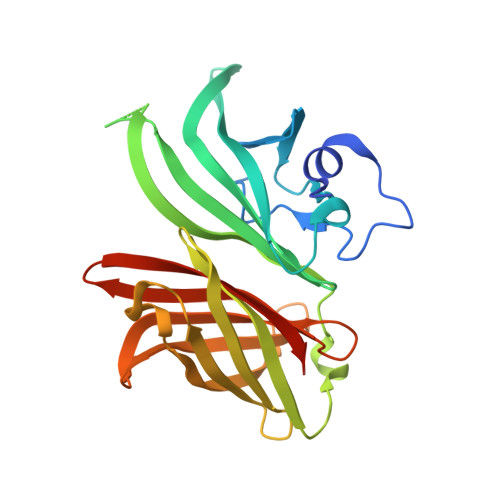
Structural characterization of a cross-protective natural chimera of factor H binding protein from meningococcal serogroup B strain NL096.
Veggi, D., Malito, E., Lo Surdo, P., Pansegrau, W., Rippa, V., Wahome, N., Savino, S., Masignani, V., Pizza, M., Bottomley, M.J.(2022) Comput Struct Biotechnol J 20: 2070-2081
- PubMed: 35601959
- DOI: https://doi.org/10.1016/j.csbj.2022.04.011
- Primary Citation of Related Structures:
7NRU - PubMed Abstract:
Invasive meningococcal disease can cause fatal sepsis and meningitis and is a global health threat. Factor H binding protein (fHbp) is a protective antigen included in the two currently available vaccines against serogroup B meningococcus (MenB). FHbp is a remarkably variable surface-exposed meningococcal virulence factor with over 1300 different amino acid sequences identified so far. Based on this variability, fHbp has been classified into three variants, two subfamilies or nine modular groups, with low degrees of cross-protective activity. Here, we report the crystal structure of a natural fHbp cross-variant chimera, named variant1-2,3.x expressed by the MenB clinical isolate NL096, at 1.2 Å resolution, the highest resolution of any fHbp structure reported to date. We combined biochemical, site-directed mutagenesis and computational biophysics studies to deeply characterize this rare chimera. We determined the structure to be composed of two adjacent domains deriving from the three variants and determined the molecular basis of its stability, ability to bind Factor H and to adopt the canonical three-dimensional fHbp structure. These studies guided the design of loss-of-function mutations with potential for even greater immunogenicity. Moreover, this study represents a further step in the understanding of the fHbp biological and immunological evolution in nature. The chimeric variant1-2,3.x fHbp protein emerges as an intriguing cross-protective immunogen and suggests that identification of such naturally occurring hybrid proteins may result in stable and cross-protective immunogens when seeking to design and develop vaccines against highly variable pathogens.
- GSK Vaccines srl, Via Fiorentina 1, Siena 53100, Italy.
Organizational Affiliation: