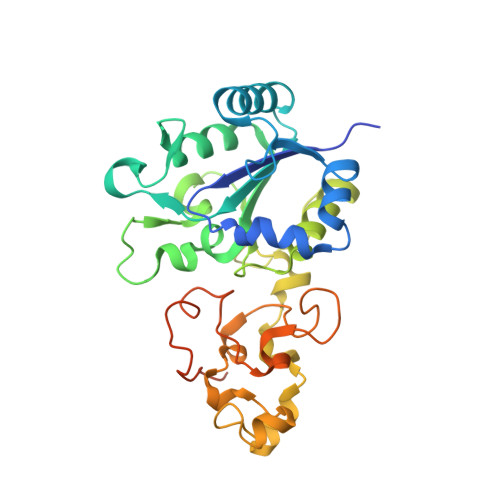
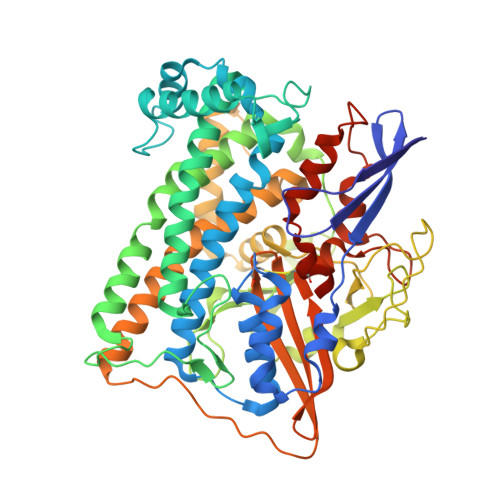
A comprehensive structural and kinetic investigation of the role of the active-site argininein bidirectional hydrogen activation by the [NiFe]-hydrogenase "Hyd-2) from Escherichia coli
Evans, R.M., Beaton, S.E., Kertiss, L., Myers, W.K., Carr, S.B., Armstrong, F.A.To be published.