Unique Pharmacological Properties of alpha-Conotoxin OmIA at alpha 7 nAChRs.
Ho, T.N.T., Abraham, N., Lewis, R.J.(2021) Front Pharmacol 12: 803397-803397
- PubMed: 34955864
- DOI: https://doi.org/10.3389/fphar.2021.803397
- Primary Citation of Related Structures:
7N43 - PubMed Abstract:
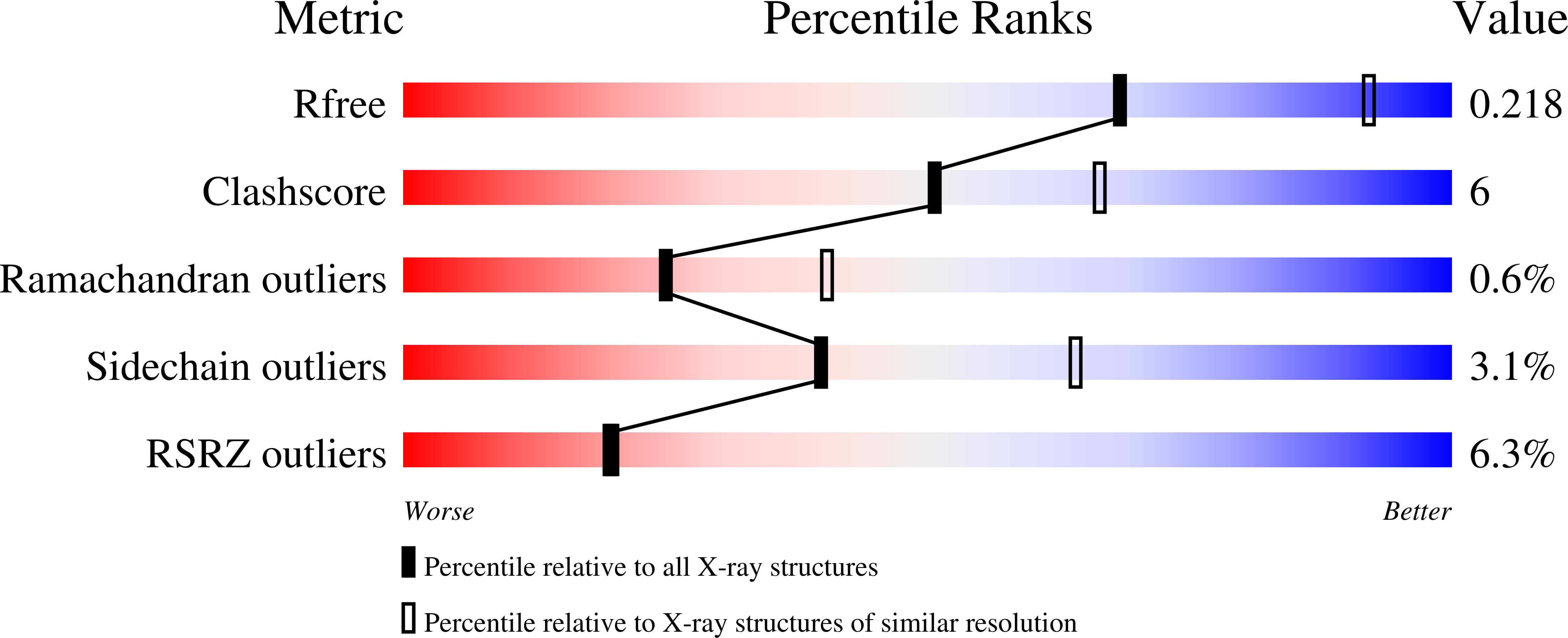
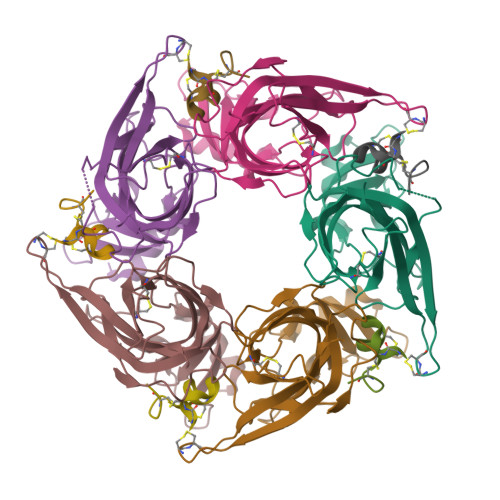
OmIA, isolated from Conus omaria venom, is a potent antagonist at α7 nAChRs. We determined the co-crystal structure of OmIA with Lymnae stagnalis acetylcholine binding protein ( Ls -AChBP) that identified His5, Val10 and Asn11 as key determinants for the high potency of OmIA at α7 nAChRs. Remarkably, despite a competitive binding mode observed in the co-crystal structure, OmIA and analogues displayed functional insurmountable antagonism at α7 and α3β4 nAChRs, except OmIA analogues having long side chain at position 10 ([V10Q]OmIA and [V10L]OmIA), which were partial insurmountable antagonist at α7 nAChRs in the presence of type II positive allosteric modulators (PAMs). A "two-state, two-step" model was used to explain these observations, with [V10Q]OmIA and [V10L]OmIA co-existing in a fast reversible/surmountable as well as a tight binding/insurmountable state. OmIA and analogues also showed biphasic-inhibition at α7 nAChRs in the presence of PNU120596, with a preference for the high-affinity binding site following prolonged exposure. The molecular basis of binding and complex pharmacological profile of OmIA at α7 nAChRs presented in here expands on the potential of α-conotoxins to probe the pharmacological properties of nAChRs and may help guide the development novel α7 modulators.
Organizational Affiliation:
Centre for Pain Research, Institute for Molecular Bioscience, The University of Queensland, St Lucia, QLD, Australia.