Structural basis of early translocation events on the ribosome.
Rundlet, E.J., Holm, M., Schacherl, M., Natchiar, S.K., Altman, R.B., Spahn, C.M.T., Myasnikov, A.G., Blanchard, S.C.(2021) Nature 595: 741-745
- PubMed: 34234344
- DOI: https://doi.org/10.1038/s41586-021-03713-x
- Primary Citation of Related Structures:
7N1P, 7N2C, 7N2U, 7N2V, 7N30, 7N31 - PubMed Abstract:
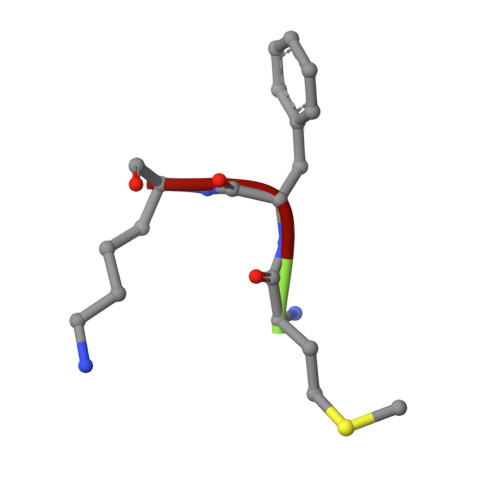
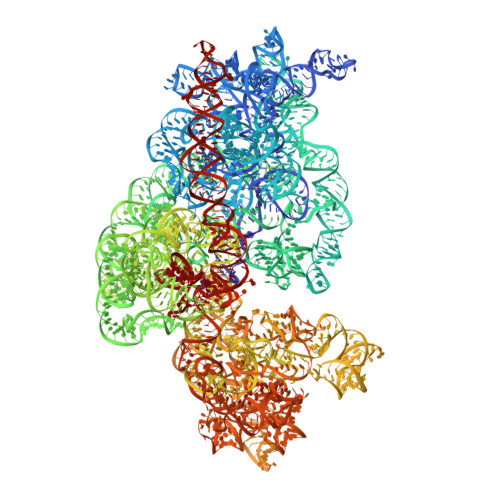
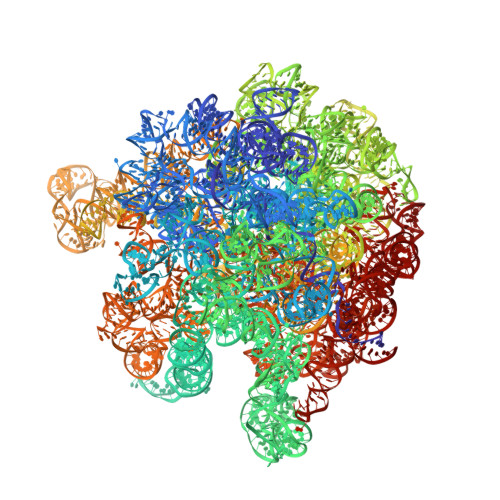
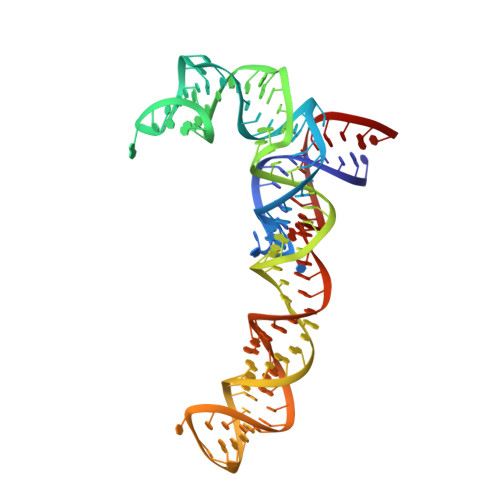
Peptide-chain elongation during protein synthesis entails sequential aminoacyl-tRNA selection and translocation reactions that proceed rapidly (2-20 per second) and with a low error rate (around 10 -3 to 10 -5 at each step) over thousands of cycles 1 . The cadence and fidelity of ribosome transit through mRNA templates in discrete codon increments is a paradigm for movement in biological systems that must hold for diverse mRNA and tRNA substrates across domains of life. Here we use single-molecule fluorescence methods to guide the capture of structures of early translocation events on the bacterial ribosome. Our findings reveal that the bacterial GTPase elongation factor G specifically engages spontaneously achieved ribosome conformations while in an active, GTP-bound conformation to unlock and initiate peptidyl-tRNA translocation. These findings suggest that processes intrinsic to the pre-translocation ribosome complex can regulate the rate of protein synthesis, and that energy expenditure is used later in the translocation mechanism than previously proposed.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, TN, USA.
Organizational Affiliation: