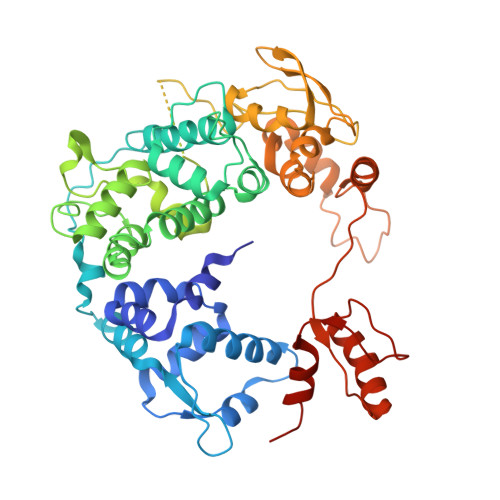
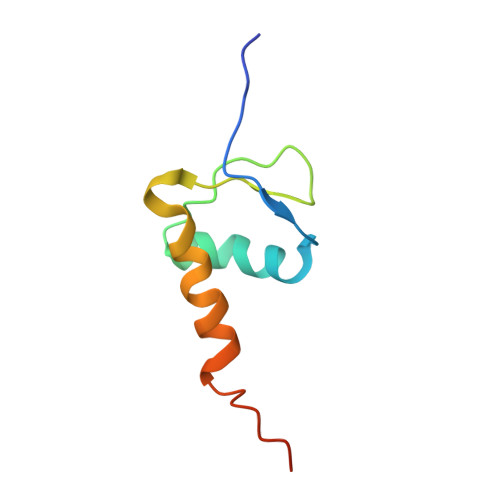
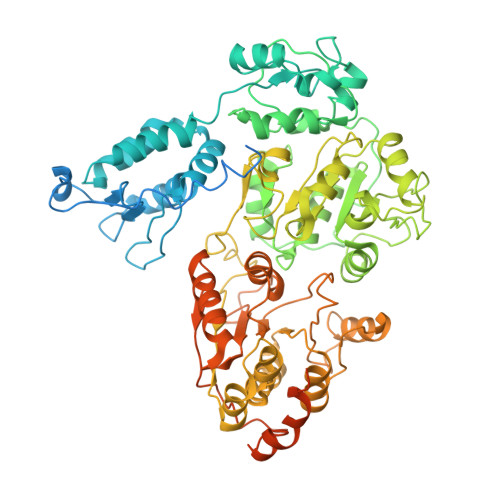
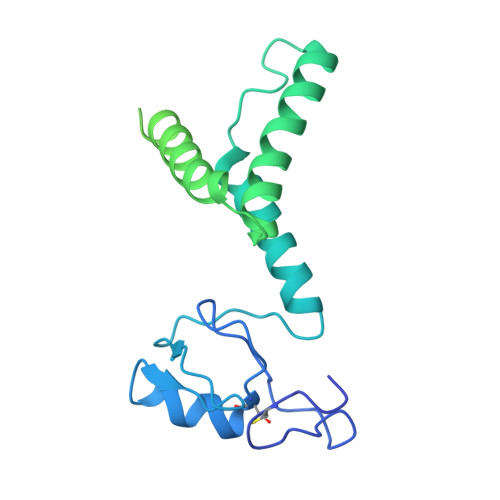
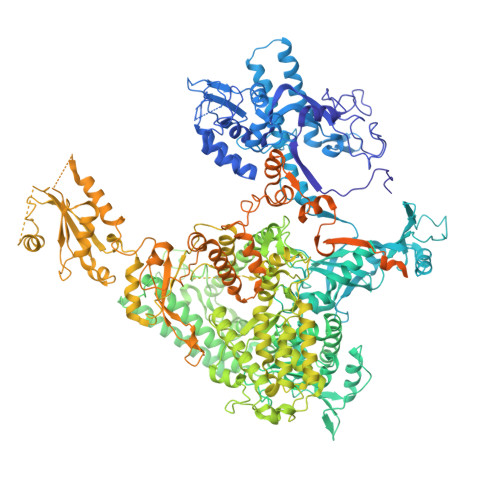
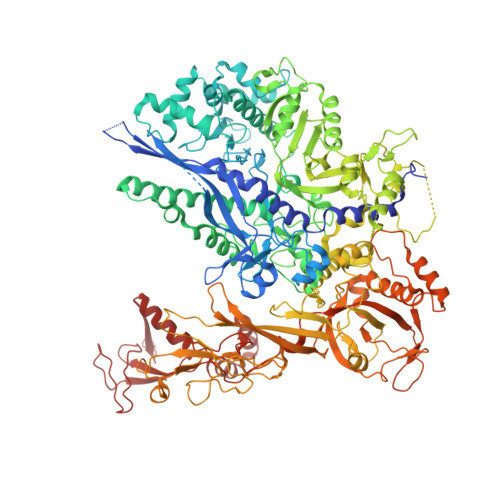
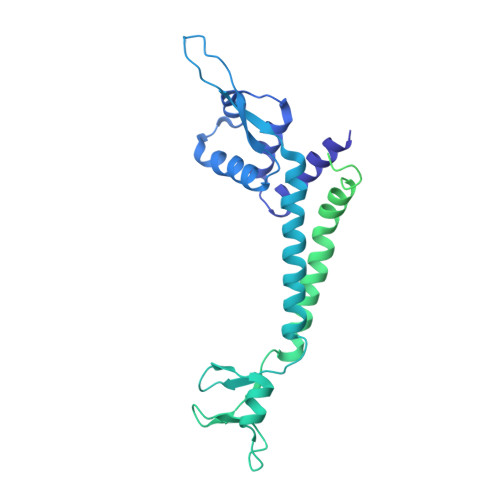
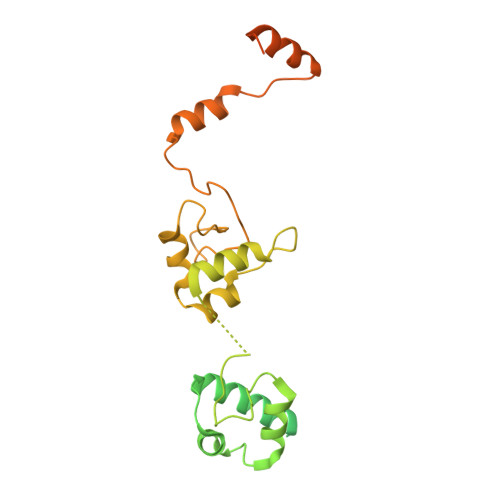
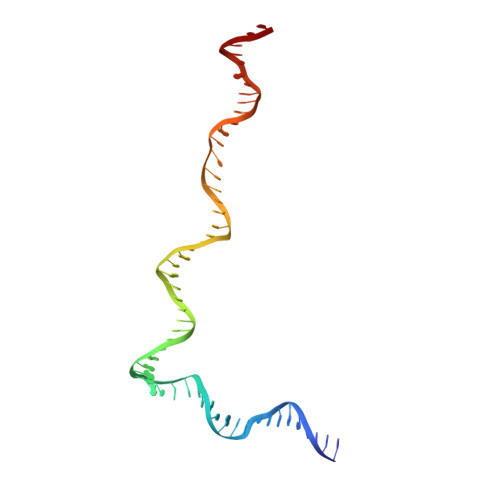
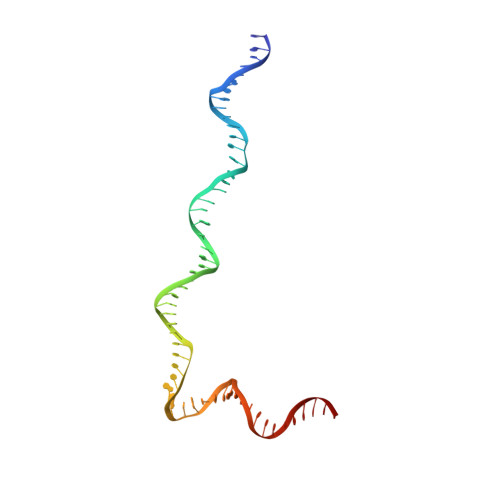
Structural visualization of de novo transcription initiation by Saccharomyces cerevisiae RNA polymerase II.
Yang, C., Fujiwara, R., Kim, H.J., Basnet, P., Zhu, Y., Gorbea Colon, J.J., Steimle, S., Garcia, B.A., Kaplan, C.D., Murakami, K.(2022) Mol Cell 82: 660-676.e9
- PubMed: 35051353
- DOI: https://doi.org/10.1016/j.molcel.2021.12.020
- Primary Citation of Related Structures:
7MEI, 7MK9, 7MKA, 7ML0, 7ML1, 7ML2, 7ML3, 7ML4 - PubMed Abstract:
Previous structural studies of the initiation-elongation transition of RNA polymerase II (pol II) transcription have relied on the use of synthetic oligonucleotides, often artificially discontinuous to capture pol II in the initiating state. Here, we report multiple structures of initiation complexes converted de novo from a 33-subunit yeast pre-initiation complex (PIC) through catalytic activities and subsequently stalled at different template positions. We determine that PICs in the initially transcribing complex (ITC) can synthesize a transcript of ∼26 nucleotides before transitioning to an elongation complex (EC) as determined by the loss of general transcription factors (GTFs). Unexpectedly, transition to an EC was greatly accelerated when an ITC encountered a downstream EC stalled at promoter proximal regions and resulted in a collided head-to-end dimeric EC complex. Our structural analysis reveals a dynamic state of TFIIH, the largest of GTFs, in PIC/ITC with distinct functional consequences at multiple steps on the pathway to elongation.
- Department of Biochemistry and Biophysics, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA 19104, USA.
Organizational Affiliation: