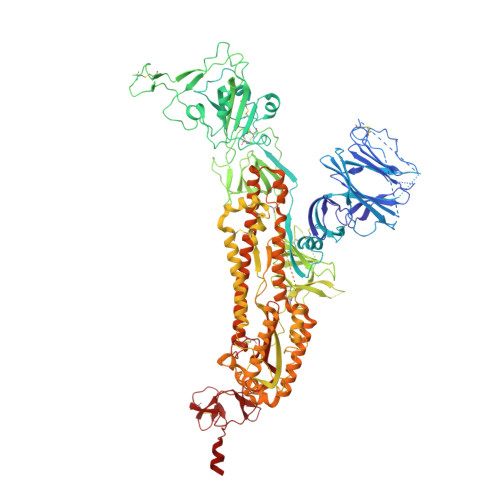
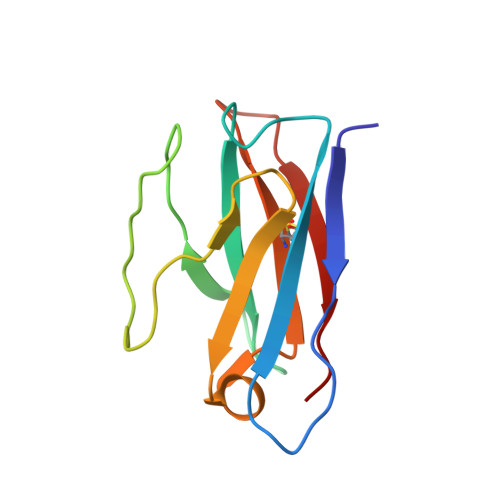
A potently neutralizing SARS-CoV-2 antibody inhibits variants of concern by utilizing unique binding residues in a highly conserved epitope.
VanBlargan, L.A., Adams, L.J., Liu, Z., Chen, R.E., Gilchuk, P., Raju, S., Smith, B.K., Zhao, H., Case, J.B., Winkler, E.S., Whitener, B.M., Droit, L., Aziati, I.D., Bricker, T.L., Joshi, A., Shi, P.Y., Creanga, A., Pegu, A., Handley, S.A., Wang, D., Boon, A.C.M., Crowe Jr., J.E., Whelan, S.P.J., Fremont, D.H., Diamond, M.S.(2021) Immunity 54: 2399-2416.e6
- PubMed: 34481543
- DOI: https://doi.org/10.1016/j.immuni.2021.08.016
- Primary Citation of Related Structures:
7MKL, 7MKM - PubMed Abstract:
With the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants with increased transmissibility and potential resistance, antibodies and vaccines with broadly inhibitory activity are needed. Here, we developed a panel of neutralizing anti-SARS-CoV-2 monoclonal antibodies (mAbs) that bound the receptor binding domain of the spike protein at distinct epitopes and blocked virus attachment to its host receptor, human angiotensin converting enzyme-2 (hACE2). Although several potently neutralizing mAbs protected K18-hACE2 transgenic mice against infection caused by ancestral SARS-CoV-2 strains, others induced escape variants in vivo or lost neutralizing activity against emerging strains. One mAb, SARS2-38, potently neutralized all tested SARS-CoV-2 variants of concern and protected mice against challenge by multiple SARS-CoV-2 strains. Structural analysis showed that SARS2-38 engaged a conserved epitope proximal to the receptor binding motif. Thus, treatment with or induction of neutralizing antibodies that bind conserved spike epitopes may limit the loss of potency of therapies or vaccines against emerging SARS-CoV-2 variants.
- Department of Medicine, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: