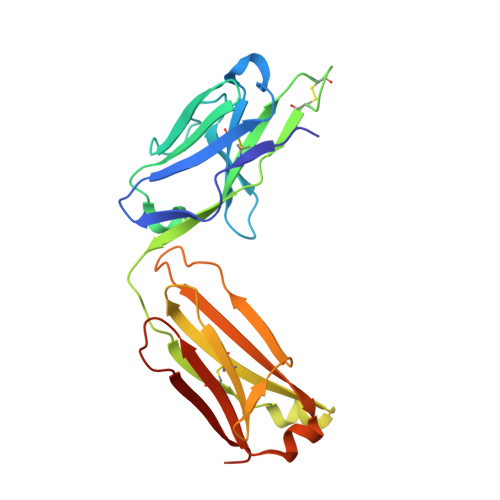
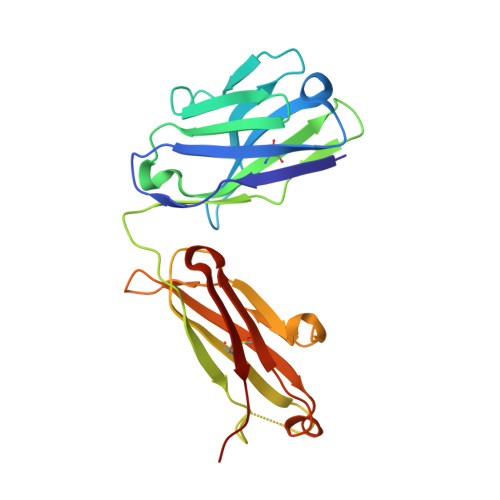
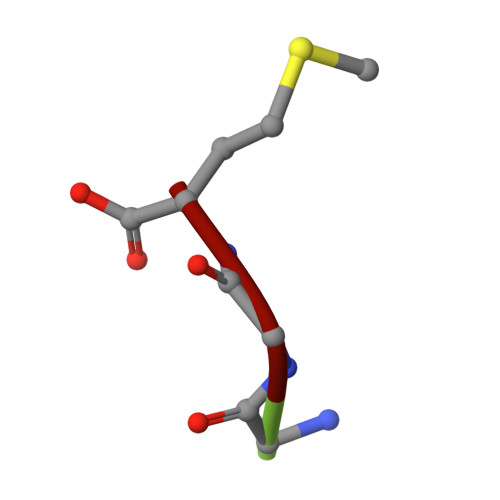
Antibody toolkit reveals N-terminally ubiquitinated substrates of UBE2W.
Davies, C.W., Vidal, S.E., Phu, L., Sudhamsu, J., Hinkle, T.B., Chan Rosenberg, S., Schumacher, F.R., Zeng, Y.J., Schwerdtfeger, C., Peterson, A.S., Lill, J.R., Rose, C.M., Shaw, A.S., Wertz, I.E., Kirkpatrick, D.S., Koerber, J.T.(2021) Nat Commun 12: 4608-4608
- PubMed: 34326324
- DOI: https://doi.org/10.1038/s41467-021-24669-6
- Primary Citation of Related Structures:
7MFR - PubMed Abstract:
The ubiquitin conjugating enzyme UBE2W catalyzes non-canonical ubiquitination on the N-termini of proteins, although its substrate repertoire remains unclear. To identify endogenous N-terminally-ubiquitinated substrates, we discover four monoclonal antibodies that selectively recognize tryptic peptides with an N-terminal diglycine remnant, corresponding to sites of N-terminal ubiquitination. Importantly, these antibodies do not recognize isopeptide-linked diglycine (ubiquitin) modifications on lysine. We solve the structure of one such antibody bound to a Gly-Gly-Met peptide to reveal the molecular basis for its selective recognition. We use these antibodies in conjunction with mass spectrometry proteomics to map N-terminal ubiquitination sites on endogenous substrates of UBE2W. These substrates include UCHL1 and UCHL5, where N-terminal ubiquitination distinctly alters deubiquitinase (DUB) activity. This work describes an antibody toolkit for enrichment and global profiling of endogenous N-terminal ubiquitination sites, while revealing functionally relevant substrates of UBE2W.
- Department of Antibody Engineering, Genentech, Inc., South San Francisco, CA, USA.
Organizational Affiliation: