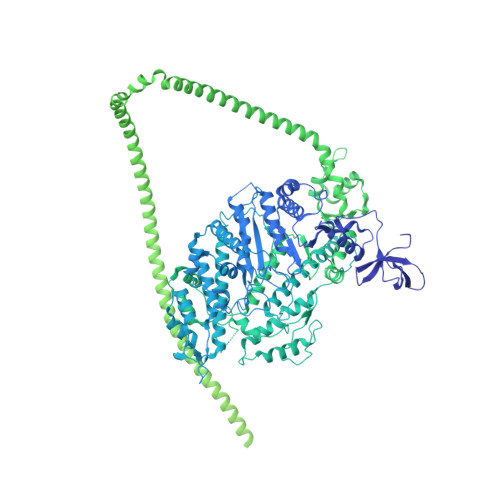
Cryo-EM structure of the autoinhibited state of myosin-2.
Heissler, S.M., Arora, A.S., Billington, N., Sellers, J.R., Chinthalapudi, K.(2021) Sci Adv 7: eabk3273-eabk3273
- PubMed: 34936462
- DOI: https://doi.org/10.1126/sciadv.abk3273
- Primary Citation of Related Structures:
7MF3 - PubMed Abstract:
We solved the near-atomic resolution structure of smooth muscle myosin-2 in the autoinhibited state (10 S ) using single-particle cryo–electron microscopy. The 3.4-Å structure reveals the precise molecular architecture of 10 S and the structural basis for myosin-2 regulation. We reveal the position of the phosphorylation sites that control myosin autoinhibition and activation by phosphorylation of the regulatory light chain. Further, we present a previously unidentified conformational state in myosin-2 that traps ADP and P i produced by the hydrolysis of ATP in the active site. This noncanonical state represents a branch of the myosin enzyme cycle and explains the autoinhibition of the enzyme function of 10 S along with its reduced affinity for actin. Together, our structure defines the molecular mechanisms that drive 10 S formation, stabilization, and relief by phosphorylation of the regulatory light chain.
- Department of Physiology and Cell Biology, Dorothy M. Davis Heart and Lung Research Institute, The Ohio State University College of Medicine, Columbus, OH, USA.
Organizational Affiliation: