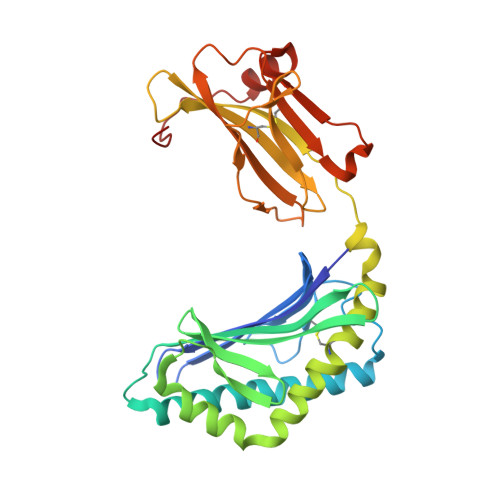
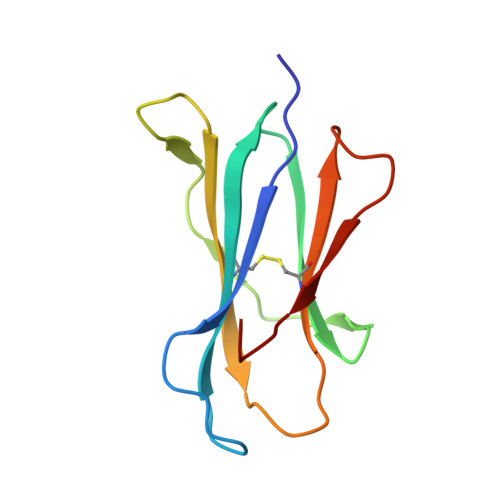
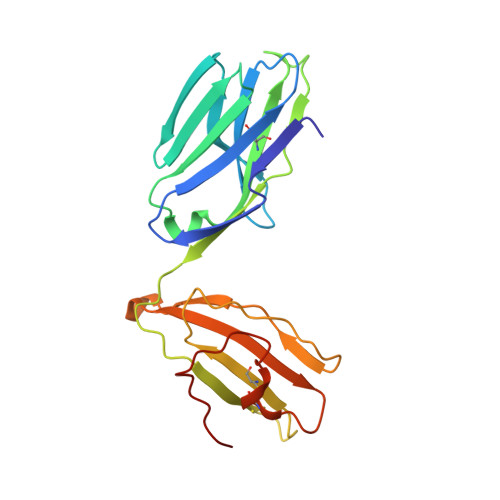
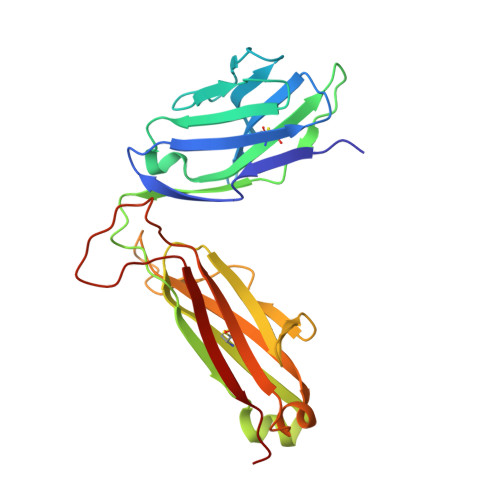
Host immunomodulatory lipids created by symbionts from dietary amino acids.
Oh, S.F., Praveena, T., Song, H., Yoo, J.S., Jung, D.J., Erturk-Hasdemir, D., Hwang, Y.S., Lee, C.C., Le Nours, J., Kim, H., Lee, J., Blumberg, R.S., Rossjohn, J., Park, S.B., Kasper, D.L.(2021) Nature 600: 302-307
- PubMed: 34759313
- DOI: https://doi.org/10.1038/s41586-021-04083-0
- Primary Citation of Related Structures:
6XNG, 7M72 - PubMed Abstract:
Small molecules derived from symbiotic microbiota critically contribute to intestinal immune maturation and regulation 1 . However, little is known about the molecular mechanisms that control immune development in the host-microbiota environment. Here, using a targeted lipidomic analysis and synthetic approach, we carried out a multifaceted investigation of immunomodulatory α-galactosylceramides from the human symbiont Bacteroides fragilis (BfaGCs). The characteristic terminal branching of BfaGCs is the result of incorporation of branched-chain amino acids taken up in the host gut by B. fragilis. A B. fragilis knockout strain that cannot metabolize branched-chain amino acids showed reduced branching in BfaGCs, and mice monocolonized with this mutant strain had impaired colonic natural killer T (NKT) cell regulation, implying structure-specific immunomodulatory activity. The sphinganine chain branching of BfaGCs is a critical determinant of NKT cell activation, which induces specific immunomodulatory gene expression signatures and effector functions. Co-crystal structure and affinity analyses of CD1d-BfaGC-NKT cell receptor complexes confirmed the interaction of BfaGCs as CD1d-restricted ligands. We present a structural and molecular-level paradigm of immunomodulatory control by interactions of endobiotic metabolites with diet, microbiota and the immune system.
- Department of Immunology, Blavatnik Institute of Harvard Medical School, Boston, MA, USA. soh2@bwh.harvard.edu.
Organizational Affiliation: