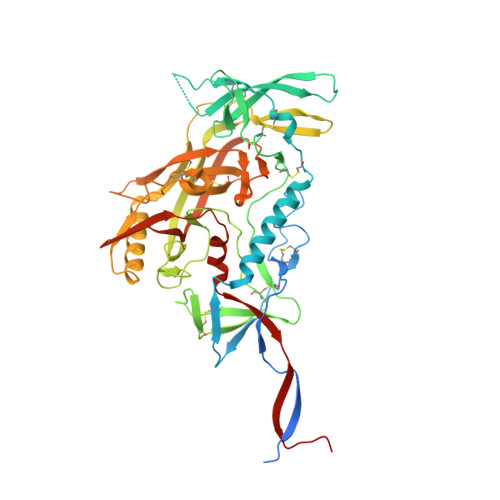
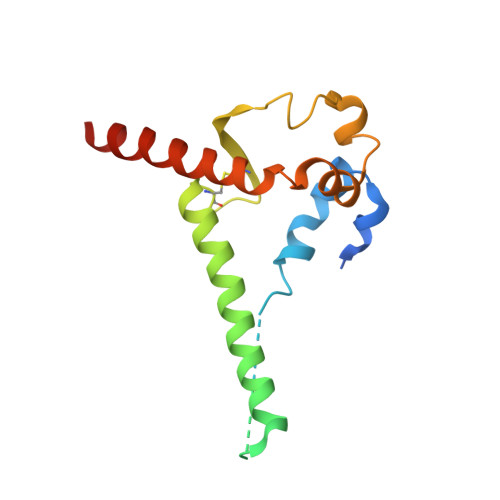
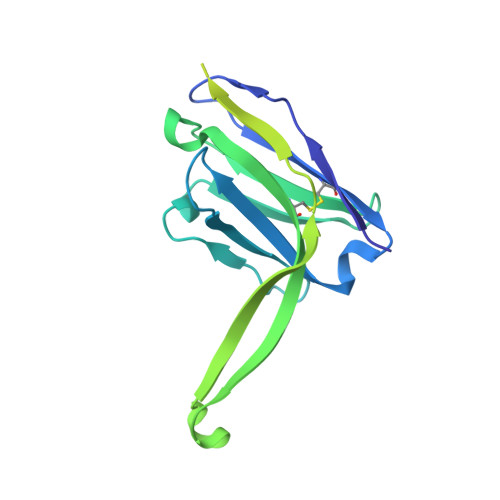
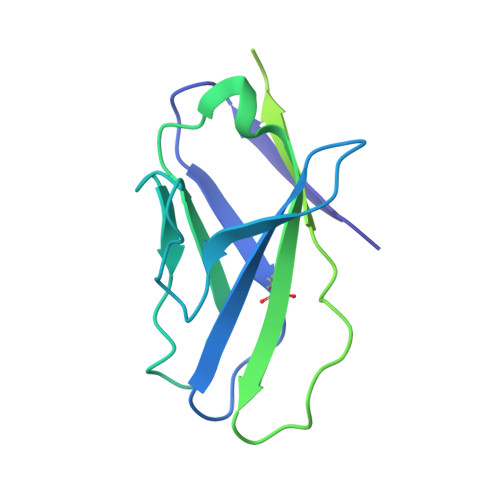
Profound structural conservation of chemically cross-linked HIV-1 envelope glycoprotein experimental vaccine antigens.
Martin, G.M., Russell, R.A., Mundsperger, P., Harris, S., Jovanoska, L., Trajano, L.F., Schiffner, T., Fabian, K., Tolazzi, M., Scarlatti, G., McFarlane, L., Cheeseman, H., Aldon, Y., Schermer, E.E., Breemen, M., Sliepen, K., Katinger, D., Kunert, R., Sanders, R.W., Shattock, R., Ward, A.B., Sattentau, Q.J.(2023) NPJ Vaccines 8: 101-101
- PubMed: 37443366
- DOI: https://doi.org/10.1038/s41541-023-00696-w
- Primary Citation of Related Structures:
7LX2, 7LX3, 7LXM, 7LXN - PubMed Abstract:
Chemical cross-linking is used to stabilize protein structures with additional benefits of pathogen and toxin inactivation for vaccine use, but its use has been restricted by the potential for local or global structural distortion. This is of particular importance when the protein in question requires a high degree of structural conservation for inducing a biological outcome such as the elicitation of antibodies to conformationally sensitive epitopes. The HIV-1 envelope glycoprotein (Env) trimer is metastable and shifts between different conformational states, complicating its use as a vaccine antigen. Here we have used the hetero-bifunctional zero-length reagent 1-Ethyl-3-(3-Dimethylaminopropyl)-Carbodiimide (EDC) to cross-link two soluble Env trimers, selected well-folded trimer species using antibody affinity, and transferred this process to good manufacturing practice (GMP) for experimental medicine use. Cross-linking enhanced trimer stability to biophysical and enzyme attack. Cryo-EM analysis revealed that cross-linking retained the overall structure with root-mean-square deviations (RMSDs) between unmodified and cross-linked Env trimers of 0.4-0.5 Å. Despite this negligible distortion of global trimer structure, we identified individual inter-subunit, intra-subunit, and intra-protomer cross-links. Antigenicity and immunogenicity of the trimers were selectively modified by cross-linking, with cross-linked ConS retaining bnAb binding more consistently than ConM. Thus, the EDC cross-linking process improves trimer stability whilst maintaining protein folding, and is readily transferred to GMP, consistent with the more general use of this approach in protein-based vaccine design.
- Department of Integrative Structural and Computational Biology, The Scripps Research Institute, La Jolla, CA, USA. gmartin@scripps.edu.
Organizational Affiliation: