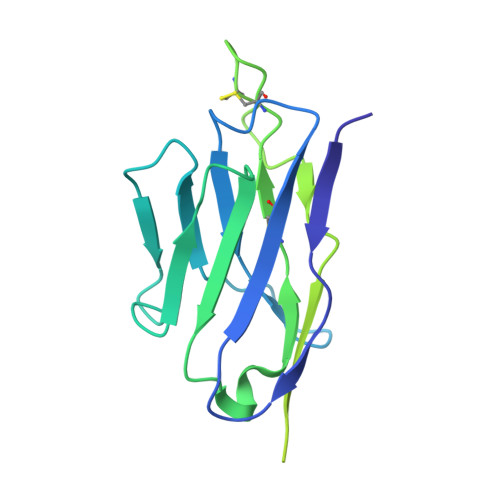
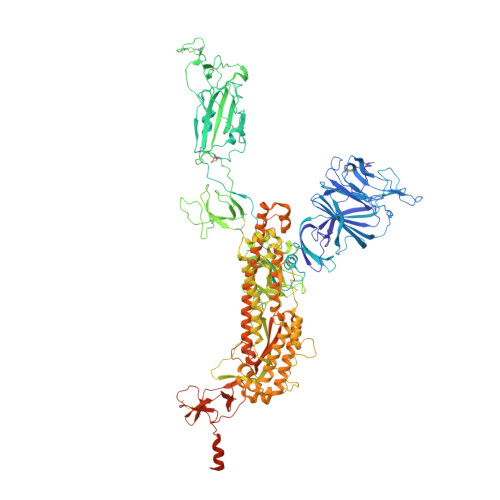
Ultrapotent antibodies against diverse and highly transmissible SARS-CoV-2 variants.
Wang, L., Zhou, T., Zhang, Y., Yang, E.S., Schramm, C.A., Shi, W., Pegu, A., Oloniniyi, O.K., Henry, A.R., Darko, S., Narpala, S.R., Hatcher, C., Martinez, D.R., Tsybovsky, Y., Phung, E., Abiona, O.M., Antia, A., Cale, E.M., Chang, L.A., Choe, M., Corbett, K.S., Davis, R.L., DiPiazza, A.T., Gordon, I.J., Hait, S.H., Hermanus, T., Kgagudi, P., Laboune, F., Leung, K., Liu, T., Mason, R.D., Nazzari, A.F., Novik, L., O'Connell, S., O'Dell, S., Olia, A.S., Schmidt, S.D., Stephens, T., Stringham, C.D., Talana, C.A., Teng, I.T., Wagner, D.A., Widge, A.T., Zhang, B., Roederer, M., Ledgerwood, J.E., Ruckwardt, T.J., Gaudinski, M.R., Moore, P.L., Doria-Rose, N.A., Baric, R.S., Graham, B.S., McDermott, A.B., Douek, D.C., Kwong, P.D., Mascola, J.R., Sullivan, N.J., Misasi, J.(2021) Science 373
- PubMed: 34210892
- DOI: https://doi.org/10.1126/science.abh1766
- Primary Citation of Related Structures:
7LRS, 7LRT, 7MLZ, 7MM0 - PubMed Abstract:
The emergence of highly transmissible SARS-CoV-2 variants of concern (VOCs) that are resistant to therapeutic antibodies highlights the need for continuing discovery of broadly reactive antibodies. We identified four receptor binding domain-targeting antibodies from three early-outbreak convalescent donors with potent neutralizing activity against 23 variants, including the B.1.1.7, B.1.351, P.1, B.1.429, B.1.526, and B.1.617 VOCs. Two antibodies are ultrapotent, with subnanomolar neutralization titers [half-maximal inhibitory concentration (IC 50 ) 0.3 to 11.1 nanograms per milliliter; IC 80 1.5 to 34.5 nanograms per milliliter). We define the structural and functional determinants of binding for all four VOC-targeting antibodies and show that combinations of two antibodies decrease the in vitro generation of escape mutants, suggesting their potential in mitigating resistance development.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: