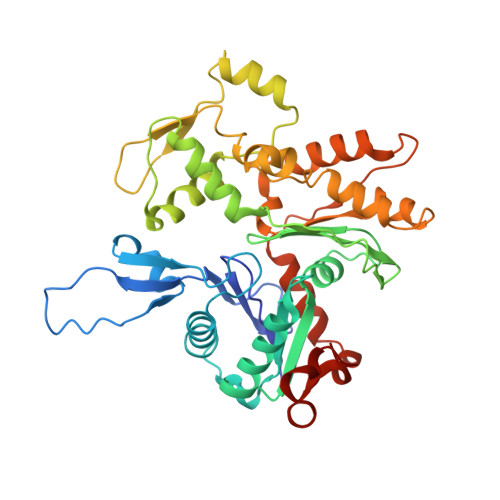
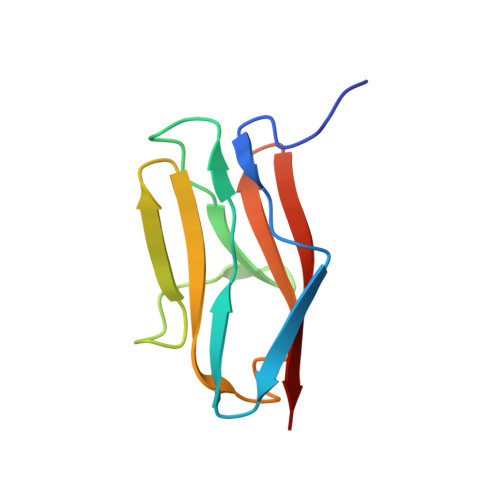
Interaction of the C2 Ig-like Domain of Cardiac Myosin Binding Protein-C with F-actin.
Risi, C.M., Patra, M., Belknap, B., Harris, S.P., White, H.D., Galkin, V.E.(2021) J Mol Biology 433: 167178-167178
- PubMed: 34329643
- DOI: https://doi.org/10.1016/j.jmb.2021.167178
- Primary Citation of Related Structures:
7LRG - PubMed Abstract:
Cardiac muscle contraction depends on interactions between thick (myosin) and thin (actin) filaments (TFs). TFs are regulated by intracellular Ca 2+ levels. Under activating conditions Ca 2+ binds to the troponin complex and displaces tropomyosin from myosin binding sites on the TF surface to allow actomyosin interactions. Recent studies have shown that in addition to Ca 2+ , the first four N-terminal domains (NTDs) of cardiac myosin binding protein C (cMyBP-C) (e.g. C0, C1, M and C2), are potent modulators of the TF activity, but the mechanism of their collective action is poorly understood. Previously, we showed that C1 activates the TF at low Ca 2+ and C0 stabilizes binding of C1 to the TF, but the ability of C2 to bind and/or affect the TF remains unknown. Here we obtained 7.5 Å resolution cryo-EM reconstruction of C2-decorated actin filaments to demonstrate that C2 binds to actin in a single structural mode that does not activate the TF unlike the polymorphic binding of C0 and C1 to actin. Comparison of amino acid sequences of C2 with either C0 or C1 shows low levels of identity between the residues involved in interactions with the TF but high levels of conservation for residues involved in Ig fold stabilization. This provides a structural basis for strikingly different interactions of structurally homologous C0, C1 and C2 with the TF. Our detailed analysis of the interaction of C2 with the actin filament provides crucial information required to model the collective action of cMyBP-C NTDs on the cardiac TF.
- Department of Physiological Sciences, Eastern Virginia Medical School, Norfolk, VA 23507, USA.
Organizational Affiliation: