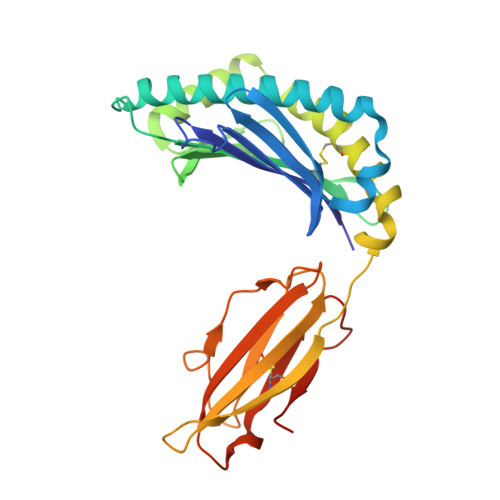
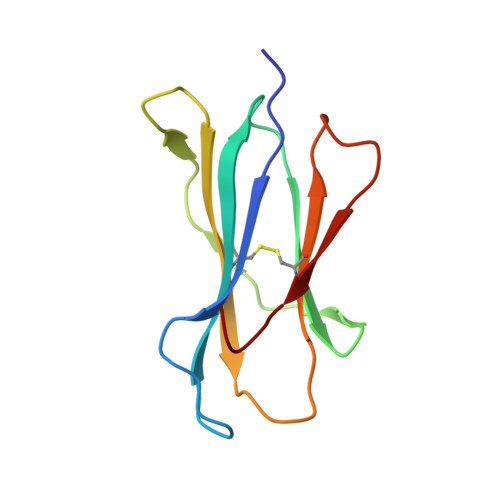
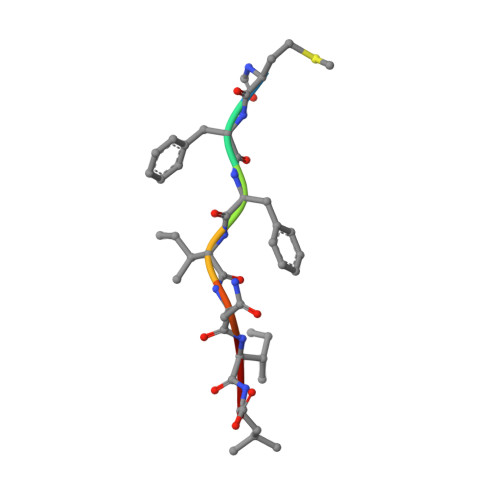
Structure and dynamics of major histocompatibility class Ib molecule H2-M3 complexed with mitochondrial-derived peptides.
Strand, A., Shen, S.T., Tomchick, D.R., Wang, J., Wang, C.R., Deisenhofer, J.(2022) J Biomol Struct Dyn 40: 10300-10312
- PubMed: 34176438
- DOI: https://doi.org/10.1080/07391102.2021.1942214
- Primary Citation of Related Structures:
7LFI, 7LFJ, 7LFK, 7LFL, 7LFM - PubMed Abstract:
Presentation of antigenic peptides to T-cell receptors is an essential step in the adaptive immune response. In the mouse the class Ib major histocompatibility complex molecule, H2-M3, presents bacterial- and mitochondrial-derived peptides to T-cell receptors on cytotoxic T cells. Four mitochondrial heptapeptides, differing only at residue 6, form complexes with H2-M3 which can be distinguished by T cells. No structures of relevant receptors are available. To investigate the structural basis for this distinction, crystal structures were determined and molecular dynamics simulations over one microsecond were done for each complex. In the crystal structures of the heptapeptide complexes with H2-M3, presented here, the side chains of the peptide residues at position 6 all point into the H2-M3 binding groove, and are thus inaccessible, so that the very similar structures do not suggest how recognition and initiation of responses by the T cells may occur. However, conformational differences, which could be crucial to T-cell discrimination, appear within one microsecond during molecular dynamics simulations of the four complexes. Specifically, the three C-terminal residues of peptide ligands with alanine or threonine at position 6 partially exit the binding groove; this does not occur in peptide ligands with isoleucine or valine at position 6. Structural changes associated with partial peptide exit from the binding groove, along with relevant peptide binding energetics and immunological results are discussed. Communicated by Ramaswamy H. Sarma.
- Green Center for Systems Biology, UT Southwestern Medical Center, Dallas, TX, USA.
Organizational Affiliation: