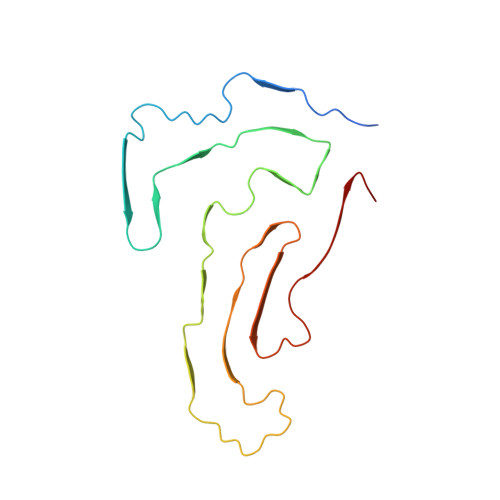
Cryo-EM structure of amyloid fibrils formed by the entire low complexity domain of TDP-43.
Li, Q., Babinchak, W.M., Surewicz, W.K.(2021) Nat Commun 12: 1620-1620
- PubMed: 33712624
- DOI: https://doi.org/10.1038/s41467-021-21912-y
- Primary Citation of Related Structures:
7KWZ - PubMed Abstract:
Amyotrophic lateral sclerosis and several other neurodegenerative diseases are associated with brain deposits of amyloid-like aggregates formed by the C-terminal fragments of TDP-43 that contain the low complexity domain of the protein. Here, we report the cryo-EM structure of amyloid formed from the entire TDP-43 low complexity domain in vitro at pH 4. This structure reveals single protofilament fibrils containing a large (139-residue), tightly packed core. While the C-terminal part of this core region is largely planar and characterized by a small proportion of hydrophobic amino acids, the N-terminal region contains numerous hydrophobic residues and has a non-planar backbone conformation, resulting in rugged surfaces of fibril ends. The structural features found in these fibrils differ from those previously found for fibrils generated from short protein fragments. The present atomic model for TDP-43 LCD fibrils provides insight into potential structural perturbations caused by phosphorylation and disease-related mutations.
- Department of Physiology and Biophysics, Case Western Reserve University, Cleveland, OH, USA.
Organizational Affiliation: