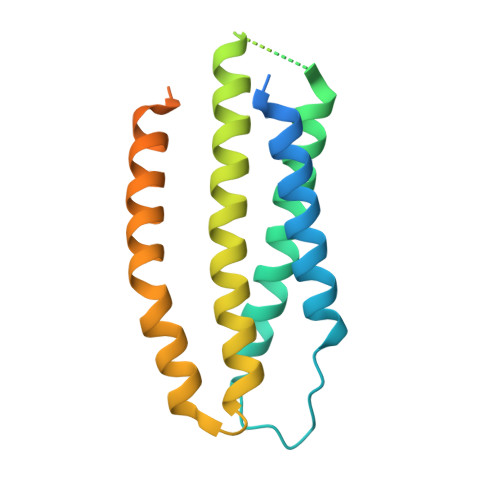
Cryo-EM structure of the calcium release-activated calcium channel Orai in an open conformation.
Hou, X., Outhwaite, I.R., Pedi, L., Long, S.B.(2020) Elife 9
- PubMed: 33252040
- DOI: https://doi.org/10.7554/eLife.62772
- Primary Citation of Related Structures:
7KR5 - PubMed Abstract:
The calcium release-activated calcium channel Orai regulates Ca 2+ entry into non-excitable cells and is required for proper immune function. While the channel typically opens following Ca 2+ release from the endoplasmic reticulum, certain pathologic mutations render the channel constitutively open. Previously, using one such mutation (H206A), we obtained low (6.7 Å) resolution X-ray structural information on Drosophila melanogaster Orai in an open conformation (Hou et al., 2018). Here we present a structure of this open conformation at 3.3 Å resolution using fiducial-assisted cryo-electron microscopy. The improved structure reveals the conformations of amino acids in the open pore, which dilates by outward movements of subunits. A ring of phenylalanine residues repositions to expose previously shielded glycine residues to the pore without significant rotational movement of the associated helices. Together with other hydrophobic amino acids, the phenylalanines act as the channel's gate. Structured M1-M2 turrets, not evident previously, form the channel's extracellular entrance.
- Structural Biology Program, Memorial Sloan Kettering Cancer Center, New York, United States.
Organizational Affiliation: