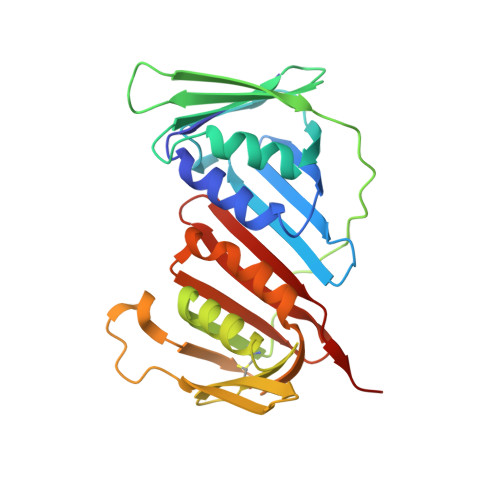
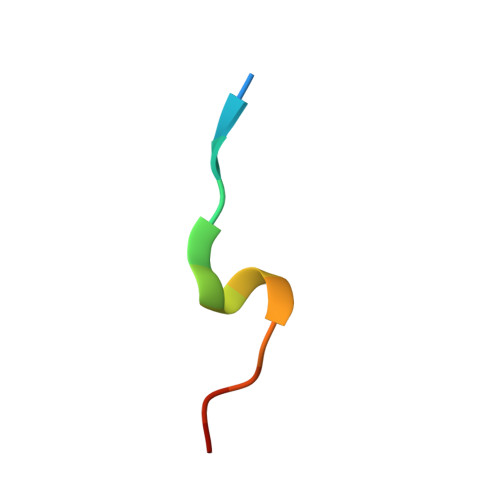
Unlocking the PIP-box: A peptide library reveals interactions that drive high-affinity binding to human PCNA.
Horsfall, A.J., Vandborg, B.A., Kowalczyk, W., Chav, T., Scanlon, D.B., Abell, A.D., Bruning, J.B.(2021) J Biological Chem 296: 100773-100773
- PubMed: 33984330
- DOI: https://doi.org/10.1016/j.jbc.2021.100773
- Primary Citation of Related Structures:
7KQ0, 7KQ1 - PubMed Abstract:
The human sliding clamp, Proliferating Cell Nuclear Antigen (hPCNA), interacts with over 200 proteins through a conserved binding motif, the PIP-box, to orchestrate DNA replication and repair. It is not clear how changes to the features of a PIP-box modulate protein binding and thus how they fine-tune downstream processes. Here, we present a systematic study of each position within the PIP-box to reveal how hPCNA-interacting peptides bind with drastically varied affinities. We synthesized a series of 27 peptides derived from the native protein p21 with small PIP-box modifications and another series of 19 peptides containing PIP-box binding motifs from other proteins. The hPCNA-binding affinity of all peptides, characterized as K D values determined by surface plasmon resonance, spanned a 4000-fold range, from 1.83 nM to 7.59 μM. The hPCNA-bound peptide structures determined by X-ray crystallography and modeled computationally revealed intermolecular and intramolecular interaction networks that correlate with high hPCNA affinity. These data informed rational design of three new PIP-box sequences, testing of which revealed the highest affinity hPCNA-binding partner to date, with a K D value of 1.12 nM, from a peptide with PIP-box QTRITEYF. This work showcases the sequence-specific nuances within the PIP-box that are responsible for high-affinity hPCNA binding, which underpins our understanding of how nature tunes hPCNA affinity to regulate DNA replication and repair processes. In addition, these insights will be useful to future design of hPCNA inhibitors.
- ARC Centre of Excellence for Nanoscale BioPhotonics, Institute of Photonics and Advanced Sensing, School of Physical Sciences, The University of Adelaide, Adelaide, South Australia, Australia.
Organizational Affiliation: