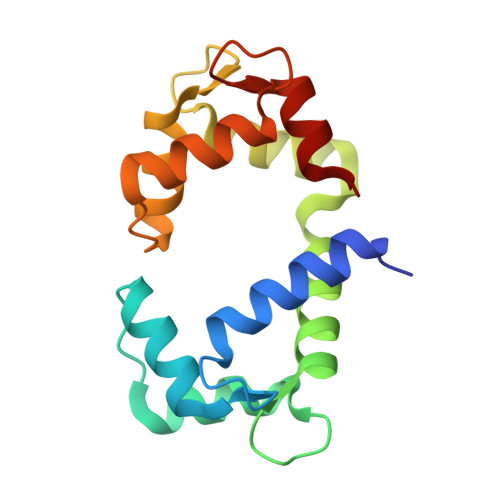
The Crystal Structure of Calmodulin Bound to the Cardiac Ryanodine Receptor (RyR2) at Residues Phe4246-Val4271 Reveals a Fifth Calcium Binding Site.
Yu, Q., Anderson, D.E., Kaur, R., Fisher, A.J., Ames, J.B.(2021) Biochemistry 60: 1088-1096
- PubMed: 33754699
- DOI: https://doi.org/10.1021/acs.biochem.1c00152
- Primary Citation of Related Structures:
7KL5 - PubMed Abstract:
Calmodulin (CaM) regulates the activity of a Ca 2+ channel known as the cardiac ryanodine receptor (RyR2), which facilitates the release of Ca 2+ from the sarcoplasmic reticulum during excitation-contraction coupling in cardiomyocytes. Mutations that disrupt this CaM-dependent channel inactivation result in cardiac arrhythmias. RyR2 contains three different CaM binding sites: CaMBD1 (residues 1940-1965), CaMBD2 (residues 3580-3611), and CaMBD3 (residues 4246-4275). Here, we report a crystal structure of Ca 2+ -bound CaM bound to RyR2 CaMBD3. The structure reveals Ca 2+ bound to the four EF-hands of CaM as well as a fifth Ca 2+ bound to CaM in the interdomain linker region involving Asp81 and Glu85. The CaM mutant E85A abolishes the binding of the fifth Ca 2+ and weakens the binding of CaMBD3 to Ca 2+ -bound CaM. Thus, the binding of the fifth Ca 2+ is important for stabilizing the complex in solution and is not a crystalline artifact. The CaMBD3 peptide in the complex adopts an α-helix (between Phe4246 and Val4271) that interacts with both lobes of CaM. Hydrophobic residues in the CaMBD3 helix (Leu4255 and Leu4259) form intermolecular contacts with the CaM N-lobe, and the CaMBD3 mutations (L4255A and L4259A) each weaken the binding of CaM to RyR2. Aromatic residues on the opposite side of the CaMBD3 helix (Phe4246 and Tyr4250) interact with the CaM C-lobe, but the mutants (F4246A and Y4250A) have no detectable effect on CaM binding in solution. We suggest that the binding of CaM to CaMBD3 and the binding of a fifth Ca 2+ to CaM may contribute to the regulation of RyR2 channel function.
- Department of Chemistry, University of California, Davis, California 95616, United States.
Organizational Affiliation: