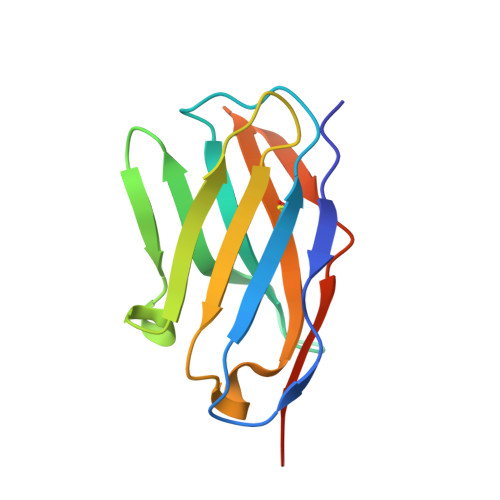
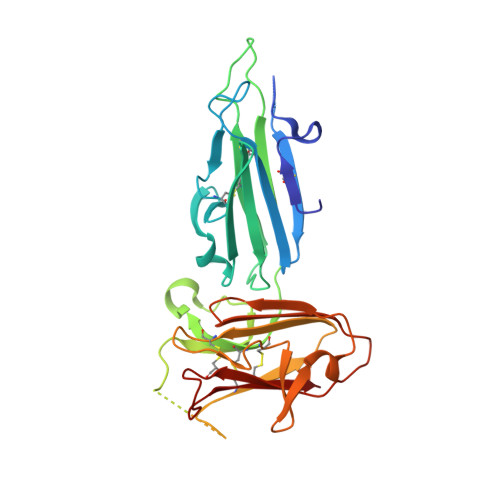
Nanobody generation and structural characterization of Plasmodium falciparum 6-cysteine protein Pf12p.
Dietrich, M.H., Chan, L.J., Adair, A., Keremane, S., Pymm, P., Lo, A.W., Cao, Y.C., Tham, W.H.(2021) Biochem J 478: 579-595
- PubMed: 33480416
- DOI: https://doi.org/10.1042/BCJ20200415
- Primary Citation of Related Structures:
7KJ7, 7KJH, 7KJI - PubMed Abstract:
Surface-associated proteins play critical roles in the Plasmodium parasite life cycle and are major targets for vaccine development. The 6-cysteine (6-cys) protein family is expressed in a stage-specific manner throughout Plasmodium falciparum life cycle and characterized by the presence of 6-cys domains, which are β-sandwich domains with conserved sets of disulfide bonds. Although several 6-cys family members have been implicated to play a role in sexual stages, mosquito transmission, evasion of the host immune response and host cell invasion, the precise function of many family members is still unknown and structural information is only available for four 6-cys proteins. Here, we present to the best of our knowledge, the first crystal structure of the 6-cys protein Pf12p determined at 2.8 Å resolution. The monomeric molecule folds into two domains, D1 and D2, both of which adopt the canonical 6-cys domain fold. Although the structural fold is similar to that of Pf12, its paralog in P. falciparum, we show that Pf12p does not complex with Pf41, which is a known interaction partner of Pf12. We generated 10 distinct Pf12p-specific nanobodies which map into two separate epitope groups; one group which binds within the D2 domain, while several members of the second group bind at the interface of the D1 and D2 domain of Pf12p. Characterization of the structural features of the 6-cys family and their associated nanobodies provide a framework for generating new tools to study the diverse functions of the 6-cys protein family in the Plasmodium life cycle.
- The Walter and Eliza Hall Institute of Medical Research, Infectious Diseases and Immune Defences Division, Parkville, Victoria, Australia.
Organizational Affiliation: