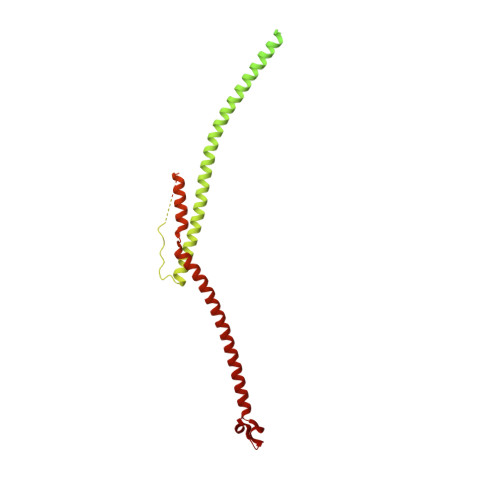
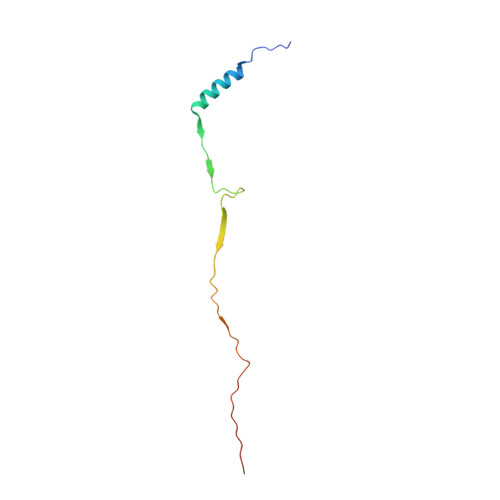
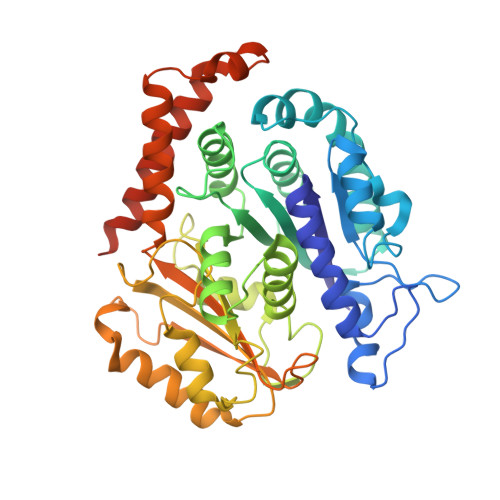
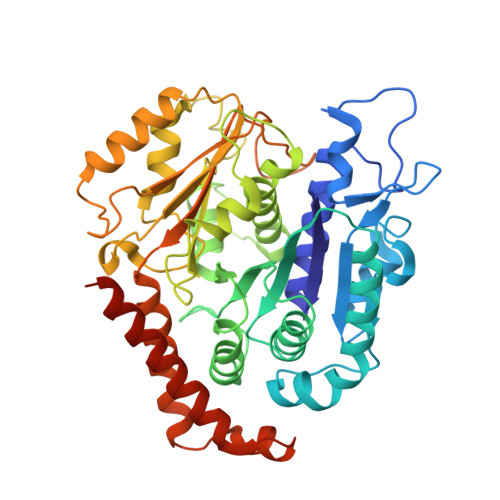
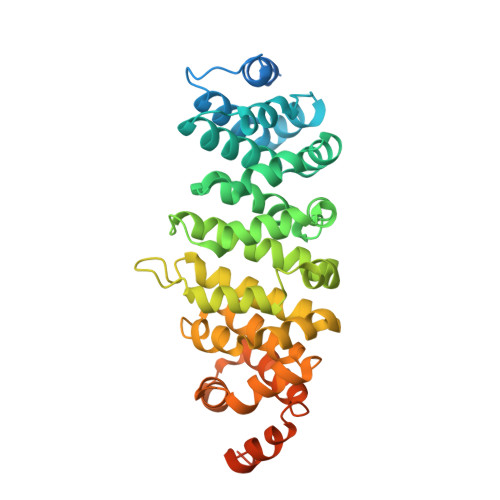
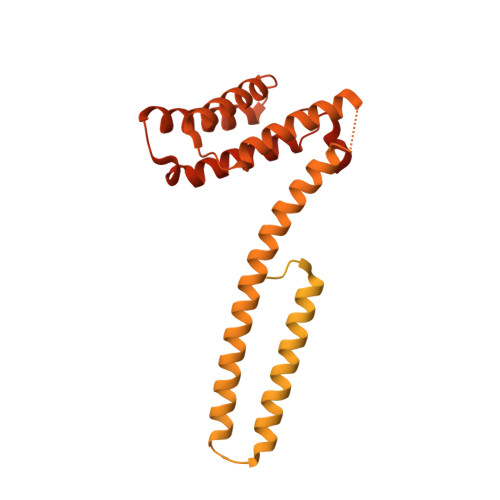
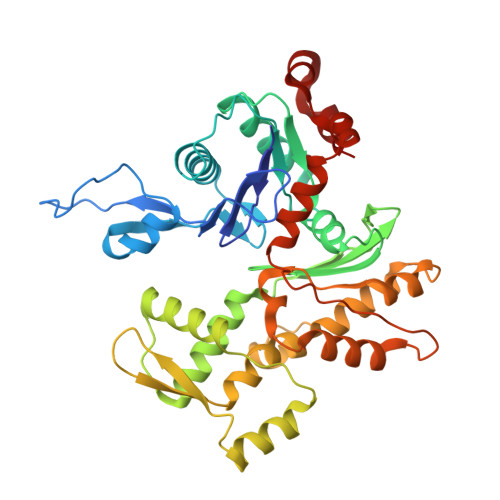
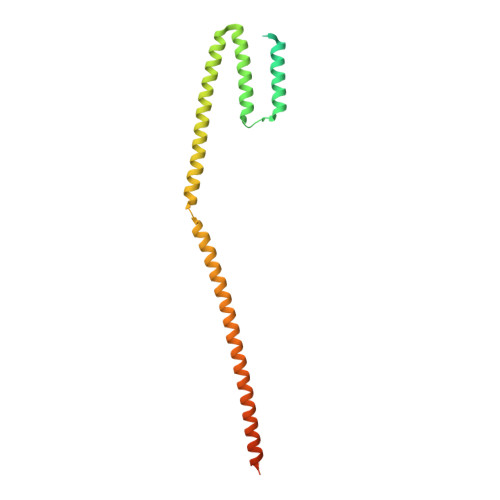
Structures of radial spokes and associated complexes important for ciliary motility.
Gui, M., Ma, M., Sze-Tu, E., Wang, X., Koh, F., Zhong, E.D., Berger, B., Davis, J.H., Dutcher, S.K., Zhang, R., Brown, A.(2021) Nat Struct Mol Biol 28: 29-37
- PubMed: 33318703
- DOI: https://doi.org/10.1038/s41594-020-00530-0
- Primary Citation of Related Structures:
7JTK, 7JTS, 7JU4 - PubMed Abstract:
In motile cilia, a mechanoregulatory network is responsible for converting the action of thousands of dynein motors bound to doublet microtubules into a single propulsive waveform. Here, we use two complementary cryo-EM strategies to determine structures of the major mechanoregulators that bind ciliary doublet microtubules in Chlamydomonas reinhardtii. We determine structures of isolated radial spoke RS1 and the microtubule-bound RS1, RS2 and the nexin-dynein regulatory complex (N-DRC). From these structures, we identify and build atomic models for 30 proteins, including 23 radial-spoke subunits. We reveal how mechanoregulatory complexes dock to doublet microtubules with regular 96-nm periodicity and communicate with one another. Additionally, we observe a direct and dynamically coupled association between RS2 and the dynein motor inner dynein arm subform c (IDAc), providing a molecular basis for the control of motor activity by mechanical signals. These structures advance our understanding of the role of mechanoregulation in defining the ciliary waveform.
- Department of Biological Chemistry and Molecular Pharmacology, Blavatnik Institute, Harvard Medical School, Boston, MA, USA.
Organizational Affiliation: