RNA stabilization by a poly(A) tail 3'-end binding pocket and other modes of poly(A)-RNA interaction.
Torabi, S.F., Vaidya, A.T., Tycowski, K.T., DeGregorio, S.J., Wang, J., Shu, M.D., Steitz, T.A., Steitz, J.A.(2021) Science 371
- PubMed: 33414189
- DOI: https://doi.org/10.1126/science.abe6523
- Primary Citation of Related Structures:
7JNH - PubMed Abstract:
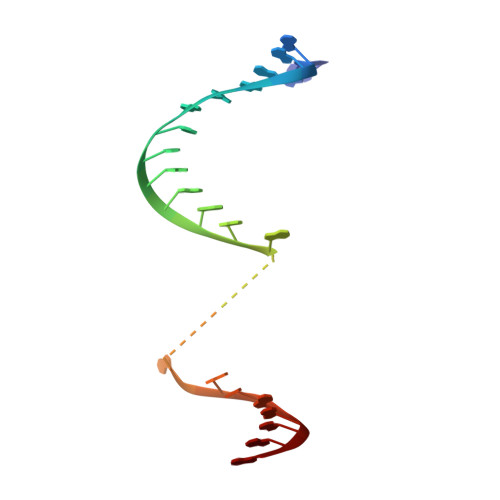
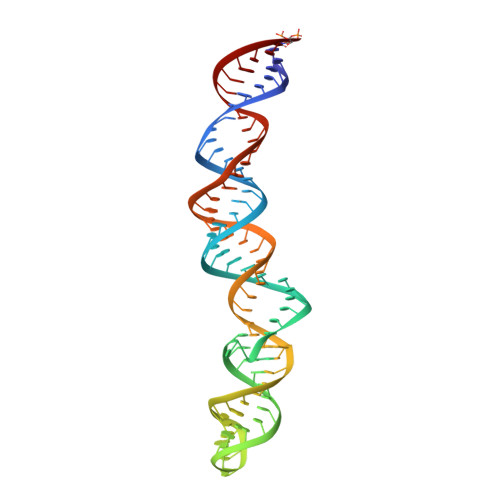
Polyadenylate [poly(A)] tail addition to the 3' end of a wide range of RNAs is a highly conserved modification that plays a central role in cellular RNA function. Elements for nuclear expression (ENEs) are cis-acting RNA elements that stabilize poly(A) tails by sequestering them in RNA triplex structures. A crystal structure of a double ENE from a rice hAT transposon messenger RNA complexed with poly(A) 28 at a resolution of 2.89 angstroms reveals multiple modes of interaction with poly(A), including major-groove triple helices, extended minor-groove interactions with RNA double helices, a quintuple-base motif that transitions poly(A) from minor-groove associations to major-groove triple helices, and a poly(A) 3'-end binding pocket. Our findings both expand the repertoire of motifs involved in long-range RNA interactions and provide insights into how polyadenylation can protect an RNA's extreme 3' end.
- Department of Molecular Biophysics and Biochemistry, Yale University School of Medicine, New Haven, CT 06536, USA.
Organizational Affiliation: