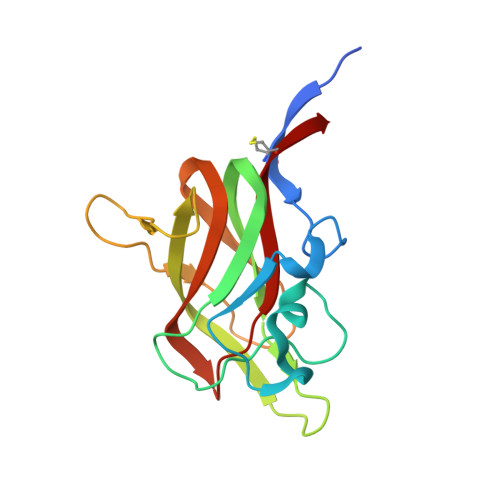
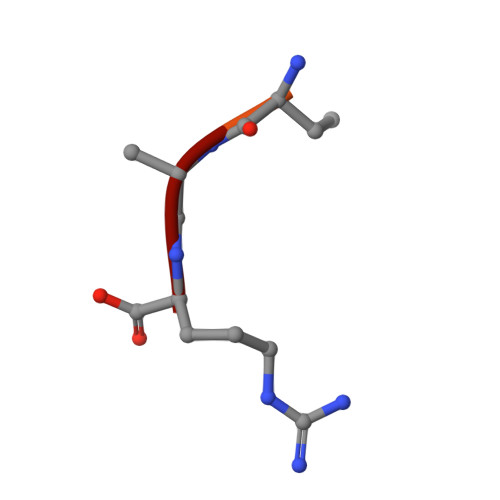
Neuropilin-1 is a host factor for SARS-CoV-2 infection.
Daly, J.L., Simonetti, B., Klein, K., Chen, K.E., Williamson, M.K., Anton-Plagaro, C., Shoemark, D.K., Simon-Gracia, L., Bauer, M., Hollandi, R., Greber, U.F., Horvath, P., Sessions, R.B., Helenius, A., Hiscox, J.A., Teesalu, T., Matthews, D.A., Davidson, A.D., Collins, B.M., Cullen, P.J., Yamauchi, Y.(2020) Science 370: 861-865
- PubMed: 33082294
- DOI: https://doi.org/10.1126/science.abd3072
- Primary Citation of Related Structures:
7JJC - PubMed Abstract:
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the causative agent of coronavirus disease 2019 (COVID-19), uses the viral spike (S) protein for host cell attachment and entry. The host protease furin cleaves the full-length precursor S glycoprotein into two associated polypeptides: S1 and S2. Cleavage of S generates a polybasic Arg-Arg-Ala-Arg carboxyl-terminal sequence on S1, which conforms to a C-end rule (CendR) motif that binds to cell surface neuropilin-1 (NRP1) and NRP2 receptors. We used x-ray crystallography and biochemical approaches to show that the S1 CendR motif directly bound NRP1. Blocking this interaction by RNA interference or selective inhibitors reduced SARS-CoV-2 entry and infectivity in cell culture. NRP1 thus serves as a host factor for SARS-CoV-2 infection and may potentially provide a therapeutic target for COVID-19.
- School of Biochemistry, Faculty of Life Sciences, Biomedical Sciences Building, University of Bristol, Bristol BS8 1TD, UK.
Organizational Affiliation: