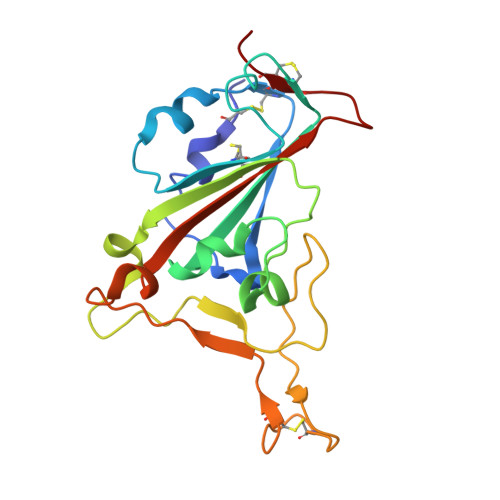
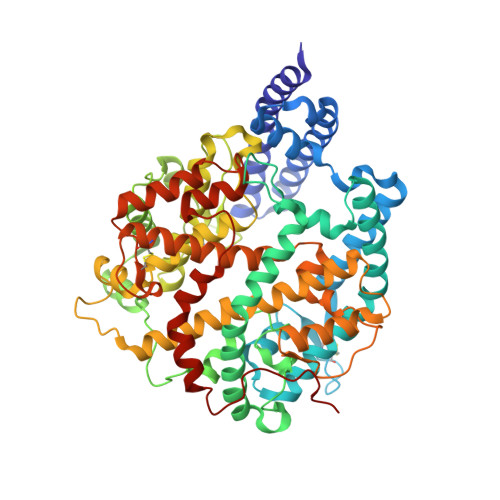
Structural insights into the binding of SARS-CoV-2, SARS-CoV, and hCoV-NL63 spike receptor-binding domain to horse ACE2.
Lan, J., Chen, P., Liu, W., Ren, W., Zhang, L., Ding, Q., Zhang, Q., Wang, X., Ge, J.(2022) Structure 30: 1432-1442.e4
- PubMed: 35917815
- DOI: https://doi.org/10.1016/j.str.2022.07.005
- Primary Citation of Related Structures:
7FC3, 7FC5, 7FC6 - PubMed Abstract:
Severe acute respiratory syndrome coronavirus (SARS-CoV), SARS-CoV-2, and human coronavirus (hCoV)-NL63 utilize ACE2 as the functional receptor for cell entry, which leads to zoonotic infection. Horses (Equus caballus) attracted our attention because the spike protein receptor-binding domains (RBDs) of SARS-CoV-2 and SARS-CoV-2-related coronaviruses bind equine ACE2 (eACE2) with high affinity. Here we show that eACE2 binds the RBDs of these three coronaviruses and also SARS-CoV-2 variants but with lower affinities compared with human ACE2 (hACE2). Structural analysis and mutation assays indicated that eACE2-H41 accounts for the lower binding affinity of eACE2 to the RBDs of SARS-CoV-2 variants (Alpha, Beta, and Gamma), SARS-CoV, and hCoV-NL63. Pseudovirus infection assays showed that the SARS-CoV-2 Delta strain (B.1.617.2) displayed a significantly increased infection efficiency in eACE2-expressing HeLa cells. Our results reveal the molecular basis of eACE2 binding to the RBDs of SARS-CoV, SARS-CoV-2, and hCoV-NL63, which provides insights into the potential animal transmission of these ACE2-dependent coronaviruses.
- The Ministry of Education Key Laboratory of Protein Science, Beijing Advanced Innovation Center for Structural Biology, Beijing Frontier Research Center for Biological Structure, School of Life Sciences, Tsinghua University, Beijing, China.
Organizational Affiliation: