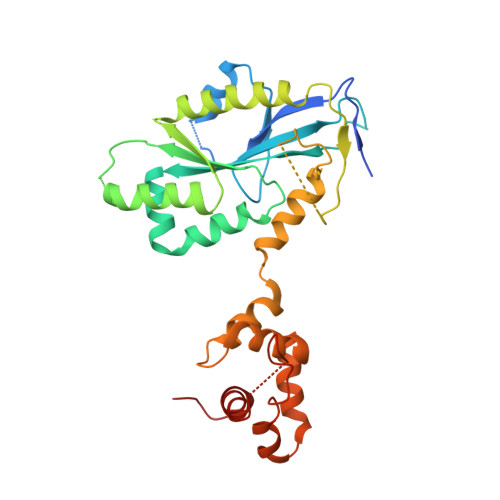
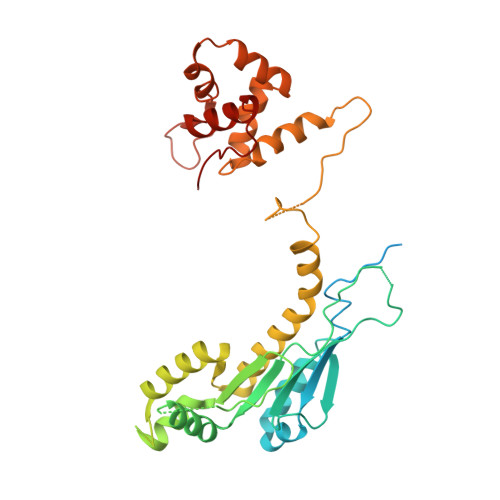
Crystal structure of the human MUS81-EME2 complex.
Hua, Z., Fang, Q., Zhang, D., Luo, Z., Yuan, C., Lin, Z.(2022) Structure 30: 743-752.e3
- PubMed: 35290797
- DOI: https://doi.org/10.1016/j.str.2022.02.015
- Primary Citation of Related Structures:
7F6L - PubMed Abstract:
MUS81 is an important structure-specific endonuclease responsible for the processing of stalled replication forks and recombination intermediates. In human, MUS81 functions by forming complexes with its regulatory subunits EME1 and EME2, playing distinct roles in G2/M and S phases. Although the structures of MUS81-EME1 have been intensively studied, there is no structure information available about MUS81-EME2. Here, we report the crystal structure of MUS81-EME2, which reveals an overall protein fold similar to that of MUS81-EME1 complex. Further biochemical and structural characterization shows that the MUS81-EME1 and MUS81-EME2 complexes are identical in substrate recognition and endonuclease activities in vitro, implying that the distinct cellular roles of the two complexes could arise from temporal controls in cells. Finally, an extensive structure-guided mutagenesis analysis provides implications for the molecular basis of how the MUS81-EME endonucleases recognize various DNA substrates in a structure-selective manner.
- College of Chemistry, Fuzhou University, Fuzhou 350108, China.
Organizational Affiliation: