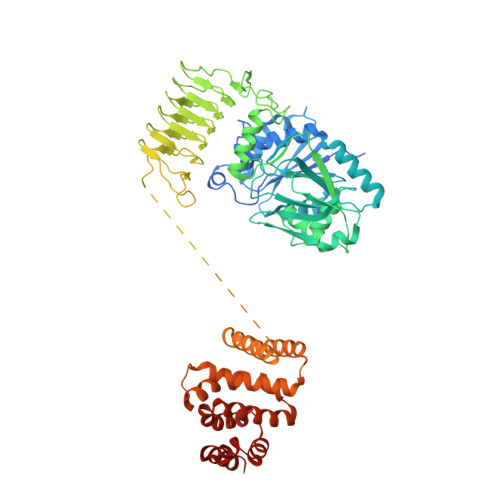
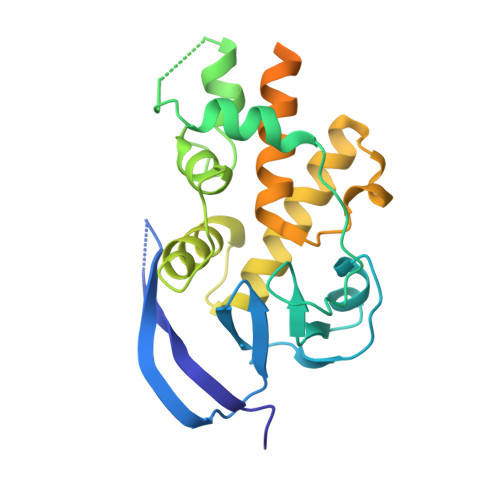
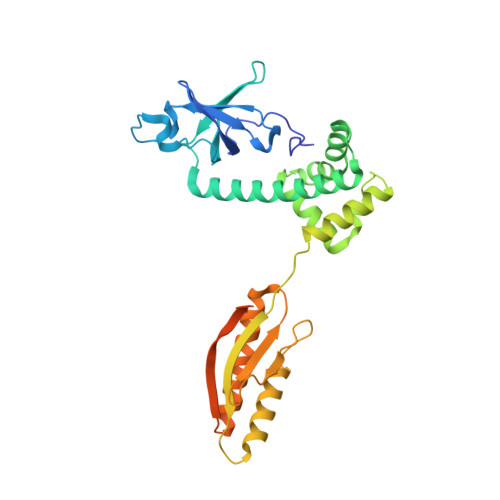
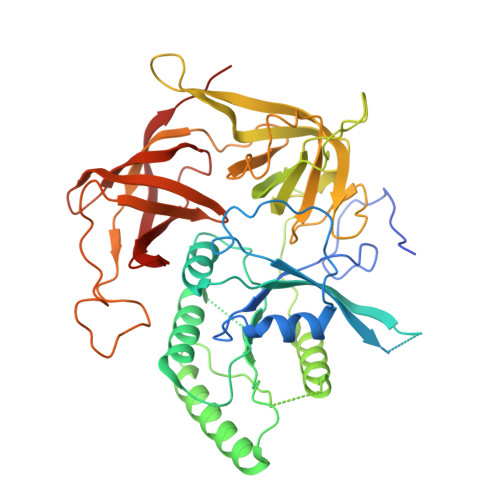
eIF2B-capturing viral protein NSs suppresses the integrated stress response.
Kashiwagi, K., Shichino, Y., Osaki, T., Sakamoto, A., Nishimoto, M., Takahashi, M., Mito, M., Weber, F., Ikeuchi, Y., Iwasaki, S., Ito, T.(2021) Nat Commun 12: 7102-7102
- PubMed: 34876589
- DOI: https://doi.org/10.1038/s41467-021-27337-x
- Primary Citation of Related Structures:
7F64, 7F66, 7F67, 7VLK - PubMed Abstract:
Various stressors such as viral infection lead to the suppression of cap-dependent translation and the activation of the integrated stress response (ISR), since the stress-induced phosphorylated eukaryotic translation initiation factor 2 [eIF2(αP)] tightly binds to eIF2B to prevent it from exchanging guanine nucleotide molecules on its substrate, unphosphorylated eIF2. Sandfly fever Sicilian virus (SFSV) evades this cap-dependent translation suppression through the interaction between its nonstructural protein NSs and host eIF2B. However, its precise mechanism has remained unclear. Here, our cryo-electron microscopy (cryo-EM) analysis reveals that SFSV NSs binds to the α-subunit of eIF2B in a competitive manner with eIF2(αP). Together with SFSV NSs, eIF2B retains nucleotide exchange activity even in the presence of eIF2(αP), in line with the cryo-EM structures of the eIF2B•SFSV NSs•unphosphorylated eIF2 complex. A genome-wide ribosome profiling analysis clarified that SFSV NSs expressed in cultured human cells attenuates the ISR triggered by thapsigargin, an endoplasmic reticulum stress inducer. Furthermore, SFSV NSs introduced in rat hippocampal neurons and human induced-pluripotent stem (iPS) cell-derived motor neurons exhibits neuroprotective effects against the ISR-inducing stress. Since ISR inhibition is beneficial in various neurological disease models, SFSV NSs may be a promising therapeutic ISR inhibitor.
- Laboratory for Translation Structural Biology, RIKEN Center for Biosystems Dynamics Research, Tsurumi-ku, Yokohama, 230-0045, Japan.
Organizational Affiliation: