Structure of CO-bound fully reduced state of bovine heart cytochrome c oxidase
Shimada, A., Shinzawa-Itoh, K., Muramoto, K., Tsukihara, T., Yoshikawa, S.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 1 | 514 | Bos taurus | Mutation(s): 0 EC: 7.1.1.9 Membrane Entity: Yes | 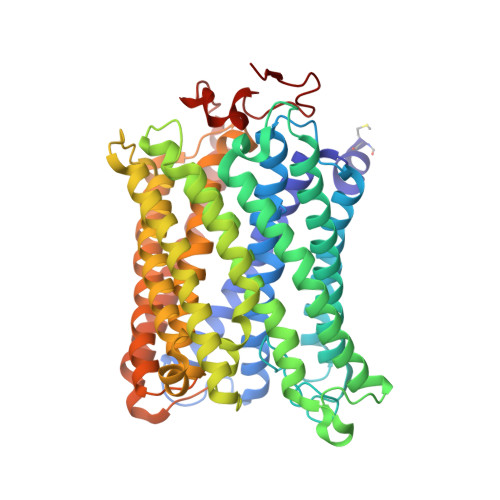 | |
UniProt | |||||
Find proteins for P00396 (Bos taurus) Explore P00396 Go to UniProtKB: P00396 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00396 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 2 | 227 | Bos taurus | Mutation(s): 0 EC: 7.1.1.9 Membrane Entity: Yes | 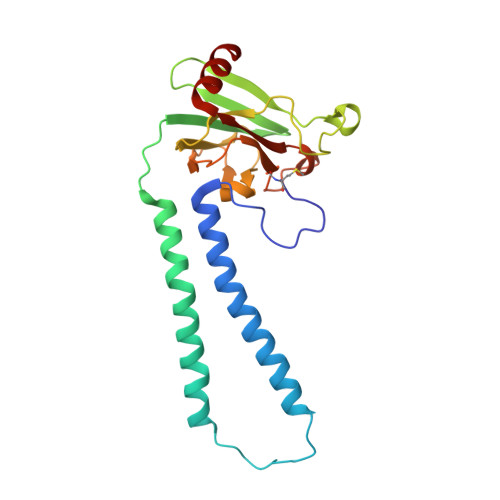 | |
UniProt | |||||
Find proteins for P68530 (Bos taurus) Explore P68530 Go to UniProtKB: P68530 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P68530 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 3 | 261 | Bos taurus | Mutation(s): 0 EC: 7.1.1.9 Membrane Entity: Yes | 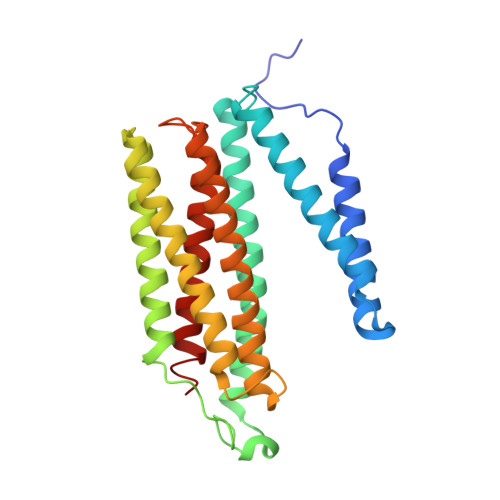 | |
UniProt | |||||
Find proteins for P00415 (Bos taurus) Explore P00415 Go to UniProtKB: P00415 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00415 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 4 isoform 1 | 147 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes | 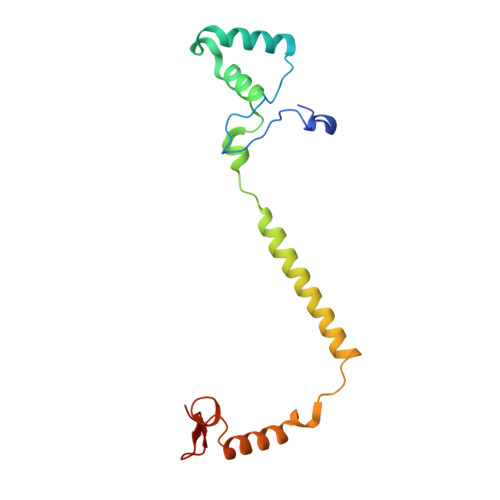 | |
UniProt | |||||
Find proteins for P00423 (Bos taurus) Explore P00423 Go to UniProtKB: P00423 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00423 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 5A | 109 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P00426 (Bos taurus) Explore P00426 Go to UniProtKB: P00426 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00426 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 6 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 5B, mitochondrial | 98 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P00428 (Bos taurus) Explore P00428 Go to UniProtKB: P00428 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00428 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 7 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 6A2 | 85 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P07471 (Bos taurus) Explore P07471 Go to UniProtKB: P07471 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07471 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 8 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 6B1 | 85 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P00429 (Bos taurus) Explore P00429 Go to UniProtKB: P00429 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00429 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 9 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 6C | 73 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P04038 (Bos taurus) Explore P04038 Go to UniProtKB: P04038 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04038 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 10 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 7A1 | 59 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P07470 (Bos taurus) Explore P07470 Go to UniProtKB: P07470 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P07470 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 11 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 7B | 56 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P13183 (Bos taurus) Explore P13183 Go to UniProtKB: P13183 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P13183 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 12 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 7C, mitochondrial | 47 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P00430 (Bos taurus) Explore P00430 Go to UniProtKB: P00430 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P00430 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 13 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Cytochrome c oxidase subunit 8B | 46 | Bos taurus | Mutation(s): 0 Membrane Entity: Yes |  | |
UniProt | |||||
Find proteins for P10175 (Bos taurus) Explore P10175 Go to UniProtKB: P10175 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P10175 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 16 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CDL (Subject of Investigation/LOI) Query on CDL | EH [auth T], TF [auth P], WB [auth C], ZC [auth G] | CARDIOLIPIN C81 H156 O17 P2 XVTUQDWPJJBEHJ-KZCWQMDCSA-L |  | ||
| TGL Query on TGL | BB [auth B] IC [auth D] IE [auth N] KG [auth Q] PH [auth Y] | TRISTEAROYLGLYCEROL C57 H110 O6 DCXXMTOCNZCJGO-UHFFFAOYSA-N |  | ||
| HEA (Subject of Investigation/LOI) Query on HEA | BA [auth A], CA [auth A], CE [auth N], DE [auth N] | HEME-A C49 H56 Fe N4 O6 ZGGYGTCPXNDTRV-PRYGPKJJSA-L |  | ||
| PEK Query on PEK | OF [auth P] PF [auth P] QF [auth P] RB [auth C] SB [auth C] | (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE C43 H78 N O8 P ANRKEHNWXKCXDB-BHFWLYLHSA-N |  | ||
| PSC Query on PSC | CB [auth B], FF [auth O] | (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE C42 H81 N O8 P JLPULHDHAOZNQI-AUSZDXHESA-O |  | ||
| PGV Query on PGV | AA [auth A] BE [auth N] HA [auth A] JE [auth N] RF [auth P] | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE C40 H77 O10 P ADYWCMPUNIVOEA-GPJPVTGXSA-N |  | ||
| DMU Query on DMU | AE [auth M] GD [auth G] HC [auth C] IG [auth P] JG [auth P] | DECYL-BETA-D-MALTOPYRANOSIDE C22 H42 O11 WOQQAWHSKSSAGF-WXFJLFHKSA-N |  | ||
| CHD Query on CHD | AD [auth G] DB [auth B] LD [auth J] LH [auth W] MF [auth P] | CHOLIC ACID C24 H40 O5 BHQCQFFYRZLCQQ-OELDTZBJSA-N |  | ||
| CUA Query on CUA | EB [auth B], GF [auth O] | DINUCLEAR COPPER ION Cu2 ALKZAGKDWUSJED-UHFFFAOYSA-N |  | ||
| PO4 Query on PO4 | IH [auth U], KD [auth H] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| ZN Query on ZN | SC [auth F], SG [auth S] | ZINC ION Zn PTFCDOFLOPIGGS-UHFFFAOYSA-N |  | ||
| CU Query on CU | DA [auth A], EE [auth N] | COPPER (II) ION Cu JPVYNHNXODAKFH-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | AB [auth A] AC [auth C] AF [auth N] AG [auth P] AH [auth S] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| CMO Query on CMO | GA [auth A], HE [auth N] | CARBON MONOXIDE C O UGFAIRIUMAVXCW-UHFFFAOYSA-N |  | ||
| MG Query on MG | EA [auth A], FE [auth N] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| NA Query on NA | FA [auth A], GE [auth N], NF [auth P], QB [auth C] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| FME Query on FME | A, N | L-PEPTIDE LINKING | C6 H11 N O3 S |  | MET |
| TPO Query on TPO | G, T | L-PEPTIDE LINKING | C4 H10 N O6 P |  | THR |
| SAC Query on SAC | I, V | L-PEPTIDE LINKING | C5 H9 N O4 |  | SER |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 181.539 | α = 90 |
| b = 203.655 | β = 90 |
| c = 177.569 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| HKL-2000 | data reduction |
| SCALEPACK | data scaling |
| MLPHARE | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Japan Society for the Promotion of Science (JSPS) | Japan | 18K06092 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 15K18493 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 18K14635 |
| Japan Society for the Promotion of Science (JSPS) | Japan | 26291033 |
| Japan Science and Technology | Japan | 12101577 |