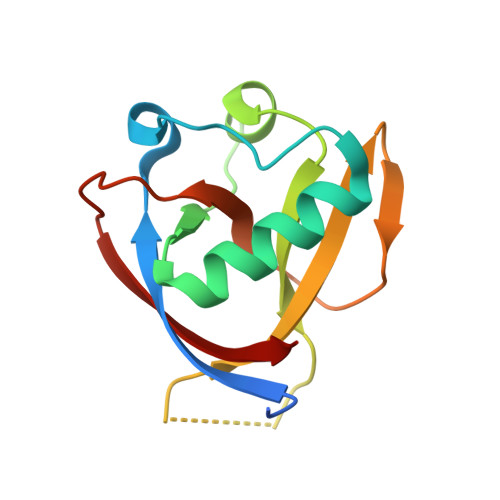
Structural Basis and Function of the N Terminus of SARS-CoV-2 Nonstructural Protein 1.
Zhao, K., Ke, Z., Hu, H., Liu, Y., Li, A., Hua, R., Guo, F., Xiao, J., Zhang, Y., Duan, L., Yan, X.F., Gao, Y.G., Liu, B., Xia, Y., Li, Y.(2021) Microbiol Spectr 9: e0016921-e0016921
- PubMed: 34132580
- DOI: https://doi.org/10.1128/Spectrum.00169-21
- Primary Citation of Related Structures:
7EQ4 - PubMed Abstract:
Nonstructural protein 1 (Nsp1) of severe acute respiratory syndrome coronaviruses (SARS-CoVs) is an important pathogenic factor that inhibits host protein translation by means of its C terminus. However, its N-terminal function remains elusive. Here, we determined the crystal structure of the N terminus (amino acids [aa] 11 to 125) of SARS-CoV-2 Nsp1 at a 1.25-Å resolution. Further functional assays showed that the N terminus of SARS-CoVs Nsp1 alone loses the ability to colocalize with ribosomes and inhibit protein translation. The C terminus of Nsp1 can colocalize with ribosomes, but its protein translation inhibition ability is significantly weakened. Interestingly, fusing the C terminus of Nsp1 with enhanced green fluorescent protein (EGFP) or other proteins in place of its N terminus restored the protein translation inhibitory ability to a level equivalent to that of full-length Nsp1. Thus, our results suggest that the N terminus of Nsp1 is able to stabilize the binding of the Nsp1 C terminus to ribosomes and act as a nonspecific barrier to block the mRNA channel, thus abrogating host mRNA translation.
- State Key Laboratory of Virology and Hubei Province Key Laboratory of Allergy and Immunology, Institute of Medical Virology, School of Basic Medical Sciences, Wuhan University, Wuhan, China.
Organizational Affiliation: