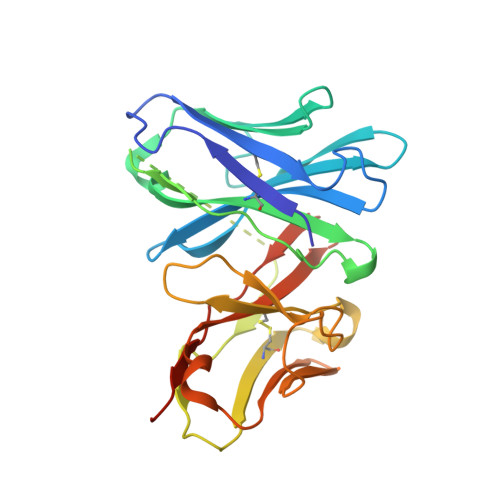
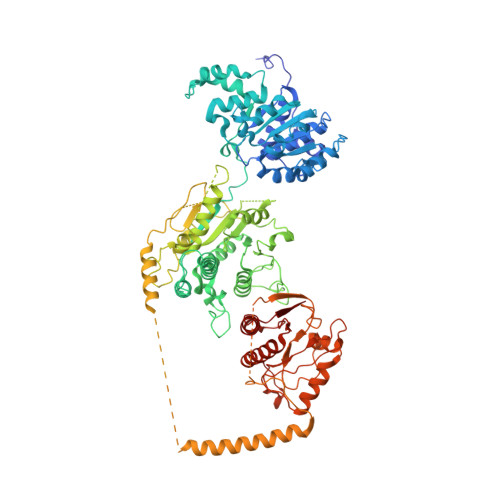
Structural basis of ALC1/CHD1L autoinhibition and the mechanism of activation by the nucleosome.
Wang, L., Chen, K., Chen, Z.(2021) Nat Commun 12: 4057-4057
- PubMed: 34210977
- DOI: https://doi.org/10.1038/s41467-021-24320-4
- Primary Citation of Related Structures:
7ENN, 7EPU - PubMed Abstract:
Chromatin remodeler ALC1 (amplification in liver cancer 1) is crucial for repairing damaged DNA. It is autoinhibited and activated by nucleosomal epitopes. However, the mechanisms by which ALC1 is regulated remain unclear. Here we report the crystal structure of human ALC1 and the cryoEM structure bound to the nucleosome. The structure shows the macro domain of ALC1 binds to lobe 2 of the ATPase motor, sequestering two elements for nucleosome recognition, explaining the autoinhibition mechanism of the enzyme. The H4 tail competes with the macro domain for lobe 2-binding, explaining the requirement for this nucleosomal epitope for ALC1 activation. A dual-arginine-anchor motif of ALC1 recognizes the acidic pocket of the nucleosome, which is critical for chromatin remodeling in vitro. Together, our findings illustrate the structures of ALC1 and shed light on its regulation mechanisms, paving the way for the discovery of drugs targeting ALC1 for the treatment of cancer.
- Key Laboratory for Protein Sciences of Ministry of Education, School of Life Science, Tsinghua University, 100084, Beijing, P.R. China.
Organizational Affiliation: