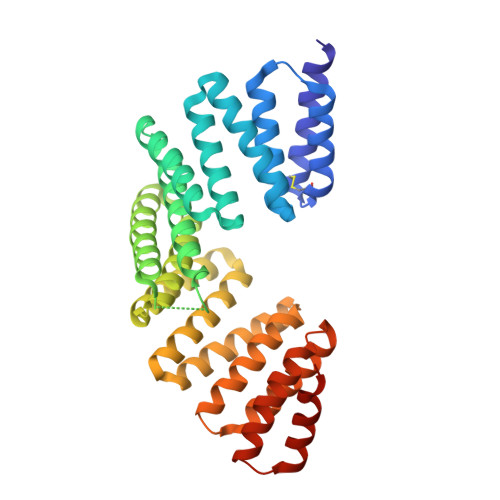
Promotion of row 1-specific tip complex condensates by Gpsm2-G alpha i provides insights into row identity of the tallest stereocilia.
Shi, Y., Lin, L., Wang, C., Zhu, J.(2022) Sci Adv 8: eabn4556-eabn4556
- PubMed: 35687681
- DOI: https://doi.org/10.1126/sciadv.abn4556
- Primary Citation of Related Structures:
7EP7 - PubMed Abstract:
The mechanosensory stereocilia in hair cells are organized into rows of graded height, a property crucial for auditory perception. Gpsm2-Gαi-Whirlin-Myo15-Eps8 complex at tips of the tallest stereocilia is proposed to define hair bundle row identity, although the underlying mechanism remains elusive. Here, we find that Gpsm2 could undergo phase separation. Moreover, row 1-specific Gpsm2-Gαi complex significantly promotes the formation of the five-component tip complex density (5xTCD) condensates. The 5xTCD condensates display much stronger actin-bundling ability than those without Gpsm2-Gαi, which may provide critical insights into how Gpsm2-Gαi specifies the tallest stereocilia. A deafness-associated mutation of Gpsm2 leads to impaired formation of the 5xTCD condensates and consequently reduced actin bundling, providing possible clues for etiology of hearing loss in patients with Chudley-McCullough syndrome.
- Department of Neurology, the First Affiliated Hospital of USTC, Ministry of Education Key Laboratory for Cellular Dynamics, Biomedical Sciences and Health Laboratory of Anhui Province, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei 230027, China.
Organizational Affiliation: