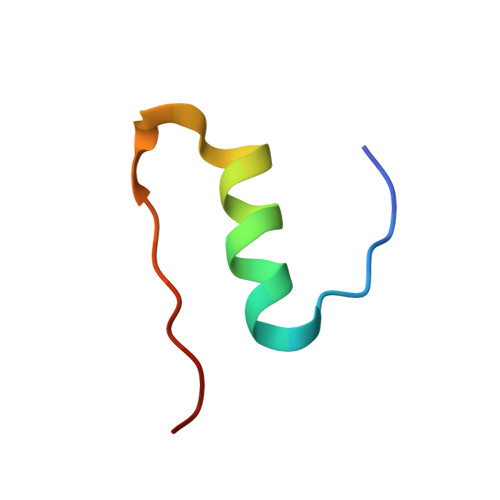
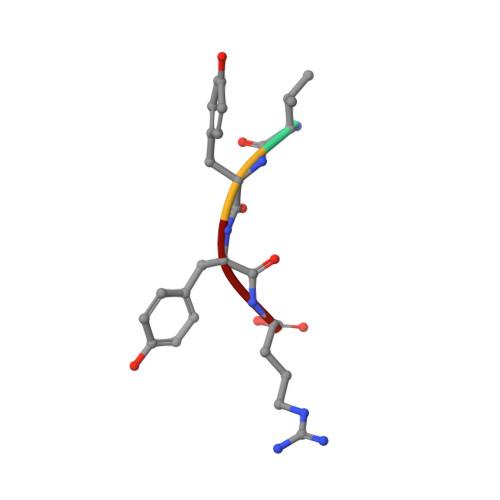
Prion-derived tetrapeptide stabilizes thermolabile insulin via conformational trapping.
Mukherjee, M., Das, D., Sarkar, J., Banerjee, N., Jana, J., Bhat, J., Reddy G, J., Bharatam, J., Chattopadhyay, S., Chatterjee, S., Chakrabarti, P.(2021) iScience 24: 102573-102573
- PubMed: 34142060
- DOI: https://doi.org/10.1016/j.isci.2021.102573
- Primary Citation of Related Structures:
7ELJ - PubMed Abstract:
Unfolding followed by fibrillation of insulin even in the presence of various excipients grappled with restricted clinical application. Thus, there is an unmet need for better thermostable, nontoxic molecules to preserve bioactive insulin under varying physiochemical perturbations. In search of cross-amyloid inhibitors, prion-derived tetrapeptide library screening reveals a consensus V(X)YR motif for potential inhibition of insulin fibrillation. A tetrapeptide VYYR, isosequential to the β2-strand of prion, effectively suppresses heat- and storage-induced insulin fibrillation and maintains insulin in a thermostable bioactive form conferring adequate glycemic control in mouse models of diabetes and impedes insulin amyloidoma formation. Besides elucidating the critical insulin-IS1 interaction (R4 of IS1 to the N24 insulin B-chain) by nuclear magnetic resonance spectroscopy, we further demonstrated non-canonical dimer-mediated conformational trapping mechanism for insulin stabilization. In this study, structural characterization and preclinical validation introduce a class of tetrapeptide toward developing thermostable therapeutically relevant insulin formulations.
- Department of Biophysics, Bose Institute, Kolkata, India.
Organizational Affiliation: