Targeting dual-specificity tyrosine phosphorylation-regulated kinase 2 with a highly selective inhibitor for the treatment of prostate cancer.
Yuan, K., Li, Z., Kuang, W., Wang, X., Ji, M., Chen, W., Ding, J., Li, J., Min, W., Sun, C., Ye, X., Lu, M., Wang, L., Ge, H., Jiang, Y., Hao, H., Xiao, Y., Yang, P.(2022) Nat Commun 13: 2903-2903
- PubMed: 35614066
- DOI: https://doi.org/10.1038/s41467-022-30581-4
- Primary Citation of Related Structures:
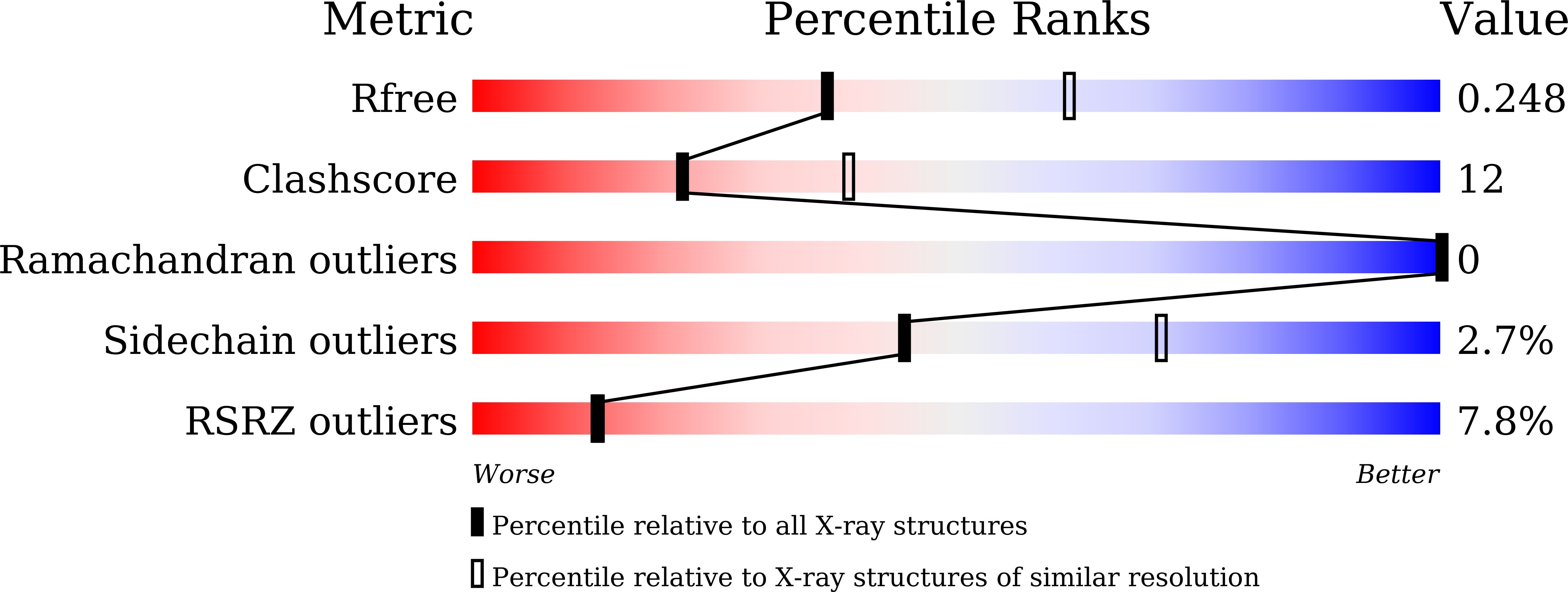
7EJV - PubMed Abstract:
Prostate cancer (PCa) is one of the most prevalent cancers in men worldwide, and hormonal therapy plays a key role in the treatment of PCa. However, the drug resistance of hormonal therapy makes it urgent and necessary to identify novel targets for PCa treatment. Herein, dual-specificity tyrosine phosphorylation-regulated kinase 2 (DYRK2) is found and confirmed to be highly expressed in the PCa tissues and cells, and knock-down of DYRK2 remarkably reduces PCa burden in vitro and in vivo. On the base of DYRK2 acting as a promising target, we further discover a highly selective DYRK2 inhibitor YK-2-69, which specifically interacts with Lys-231 and Lys-234 in the co-crystal structure. Especially, YK-2-69 exhibits more potent anti-PCa efficacy than the first-line drug enzalutamide in vivo. Meanwhile, YK-2-69 displays favorable safety properties with a maximal tolerable dose of more than 10,000 mg/kg and pharmacokinetic profiles with 56% bioavailability. In summary, we identify DYRK2 as a potential drug target and verify its critical roles in PCa. Meanwhile, we discover a highly selective DYRK2 inhibitor with favorable druggability for the treatment of PCa.
Organizational Affiliation:
State Key Laboratory of Natural Medicines and Jiangsu Key Laboratory of Drug Design and Optimization, China Pharmaceutical University, 210009, Nanjing, China.