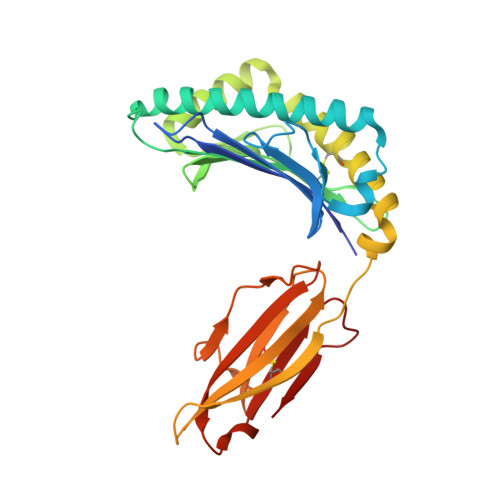
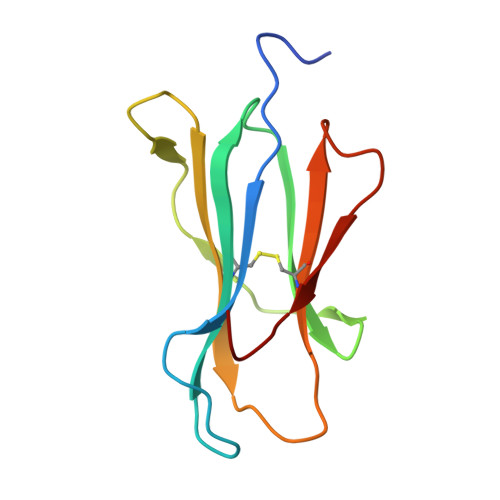
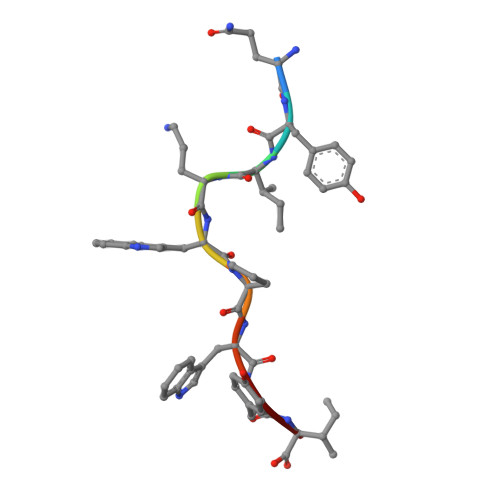
Identification of TCR repertoires in functionally competent cytotoxic T cells cross-reactive to SARS-CoV-2.
Shimizu, K., Iyoda, T., Sanpei, A., Nakazato, H., Okada, M., Ueda, S., Kato-Murayama, M., Murayama, K., Shirouzu, M., Harada, N., Hidaka, M., Fujii, S.I.(2021) Commun Biol 4: 1365-1365
- PubMed: 34857854
- DOI: https://doi.org/10.1038/s42003-021-02885-6
- Primary Citation of Related Structures:
7EJL, 7EJM, 7EJN - PubMed Abstract:
SARS-CoV-2-specific CD8 + T cells are scarce but detectable in unexposed healthy donors (UHDs). It remains unclear whether pre-existing human coronavirus (HCoV)-specific CD8 + T cells are converted to functionally competent T cells cross-reactive to SARS-CoV-2. Here, we identified the HLA-A24-high binding, immunodominant epitopes in SARS-CoV-2 spike region that can be recognized by seasonal coronavirus-specific CD8 + T cells from HLA-A24 + UHDs. Cross-reactive CD8 + T cells were clearly reduced in patients with hematological malignancy, who are usually immunosuppressed, compared to those in UHDs. Furthermore, we showed that CD8 + T cells in response to a selected dominant epitope display multifunctionality and cross-functionality across HCoVs in HLA-A24 + donors. Cross-reactivity of T-cell receptors isolated from them exhibited selective diversity at the single-cell level. Taken together, when stimulated well by immunodominant epitopes, selective pre-existing CD8 + T cells with high functional avidity may be cross-reactive against SARS-CoV-2.
- Laboratory for Immunotherapy, RIKEN Center for Integrative Medical Science (IMS), Yokohama, Japan.
Organizational Affiliation: