Structural and biochemical insights into the substrate-binding mechanism of a glycoside hydrolase family 12 beta-1,3-1,4-glucanase from Chaetomium sp.
Ma, J., Li, Y., Han, S., Jiang, Z., Yan, Q., Yang, S.(2021) J Struct Biol 213: 107774-107774
- PubMed: 34329700
- DOI: https://doi.org/10.1016/j.jsb.2021.107774
- Primary Citation of Related Structures:
7EE2, 7EEE, 7EEJ - PubMed Abstract:
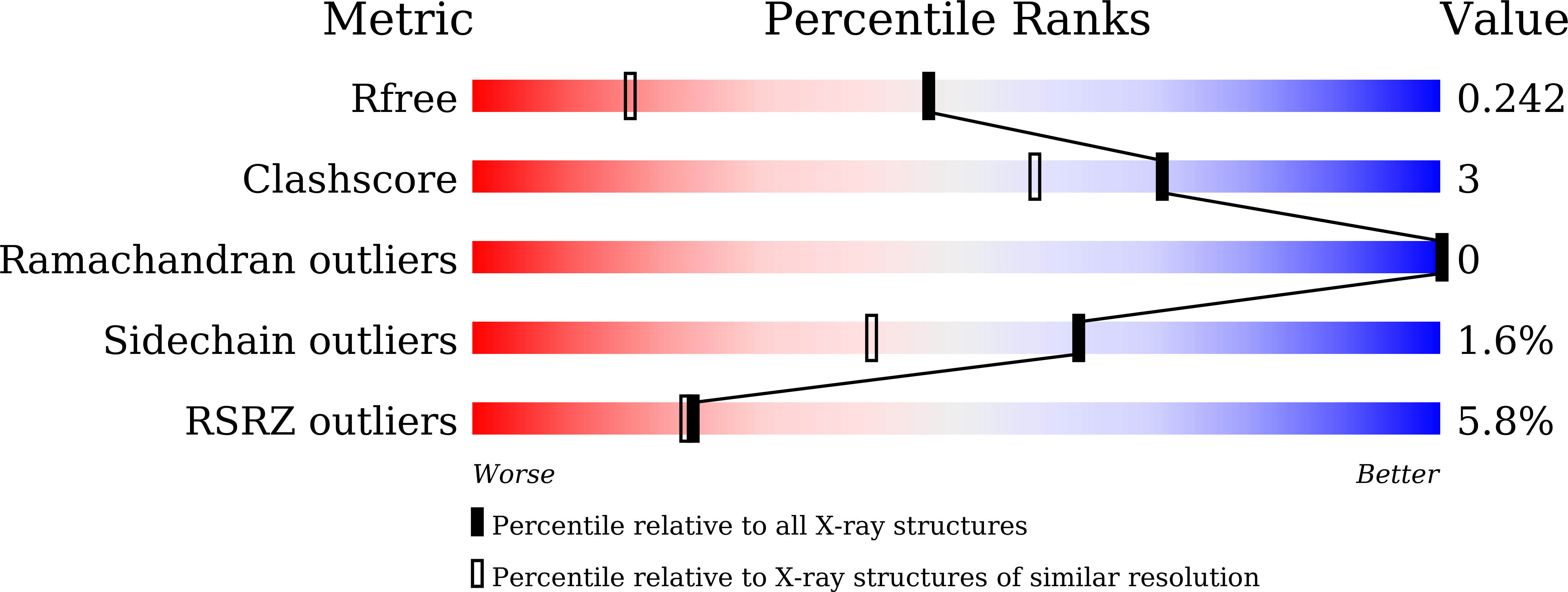
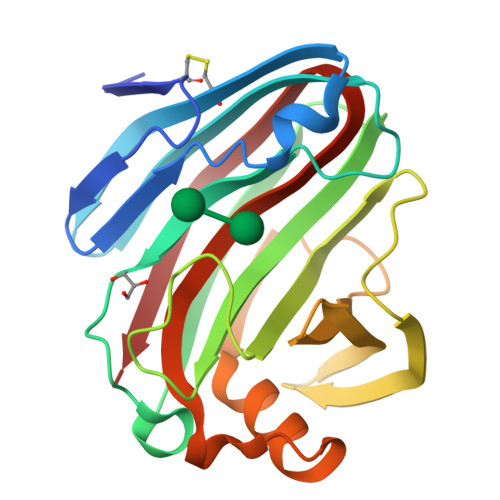
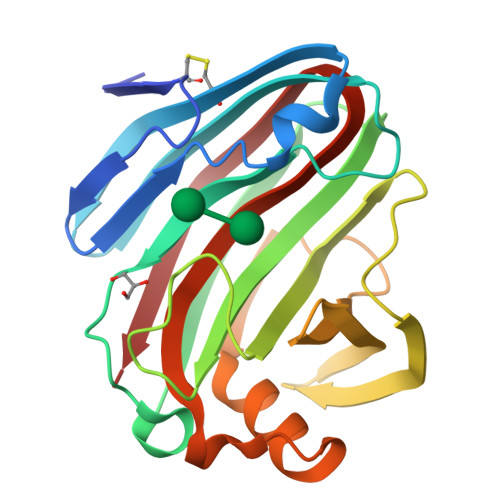
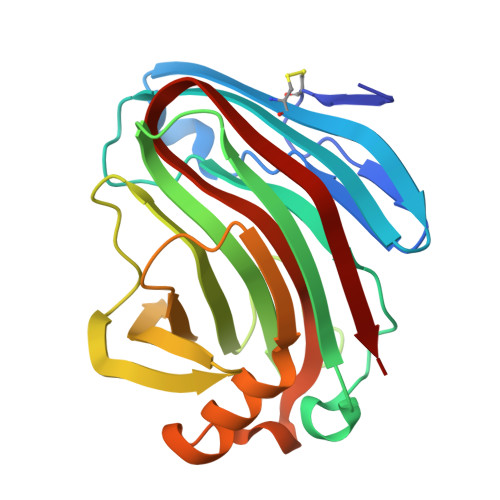
β-1,3-1,4-Glucanases are a type of hydrolytic enzymes capable of catalyzing the strict cleavage of β-1,4 glycosidic bonds adjacent to β-1,3 linkages in β-D-glucans and have exhibited great potential in food and feed industrials. In this study, a novel glycoside hydrolase (GH) family 12 β-1,3-1,4-glucanase (CtGlu12A) from the thermophilic fungus Chaetomium sp. CQ31 was identified and biochemically characterized. CtGlu12A was most active at pH 7.5 and 65 °C, respectively, and exhibited a high specific activity of 999.9 U mg -1 towards lichenin. It maintained more than 80% of its initial activity in a wide pH range of 5.0-11.0, and up to 60 °C after incubation at 55 °C for 60 min. Moreover, the crystal structures of CtGlu12A with gentiobiose and tetrasccharide were resolved. CtGlu12A had a β-jellyroll fold, and performed retaining mechanism with two glutamic acids severing as the catalytic residues. In the complex structure, cellobiose molecule showed two binding modes, occupying subsites -2 to -1 and subsites + 1 to + 2, respectively. The concave cleft made mixed β-1,3-1,4-glucan substrates maintain a bent conformation to fit into the active site. Overall, this study is not only helpful for the understanding of the substrate-binding model and catalytic mechanism of GH 12 β-1,3-1,4-glucanases, but also provides a basis for further enzymatic engineering of β-1,3-1,4-glucanases.
Organizational Affiliation:
Key Laboratory of Food Bioengineering (China National Light Industry), College of Engineering, China Agricultural University, Beijing 100083, China.