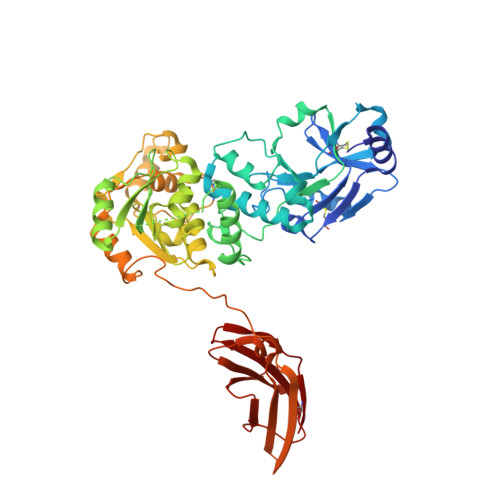
The structure of POMGNT2 provides new insights into the mechanism to determine the functional O-mannosylation site on alpha-dystroglycan.
Imae, R., Kuwabara, N., Manya, H., Tanaka, T., Tsuyuguchi, M., Mizuno, M., Endo, T., Kato, R.(2021) Genes Cells 26: 485-494
- PubMed: 33893702
- DOI: https://doi.org/10.1111/gtc.12853
- Primary Citation of Related Structures:
7E9J, 7E9K, 7E9L - PubMed Abstract:
Defects in the O-mannosyl glycan of α-dystroglycan (α-DG) are associated with α-dystroglycanopathy, a group of congenital muscular dystrophies. While α-DG has many O-mannosylation sites, only the specific positions can be modified with the functional O-mannosyl glycan, namely, core M3-type glycan. POMGNT2 is a glycosyltransferase which adds β1,4-linked GlcNAc to the O-mannose (Man) residue to acquire core M3-type glycan. Although it is assumed that POMGNT2 extends the specific O-Man residues around particular amino acid sequences, the details are not well understood. Here, we determined a series of crystal structures of POMGNT2 with and without the acceptor O-mannosyl peptides and identified the critical interactions between POMGNT2 and the acceptor peptide. POMGNT2 has an N-terminal catalytic domain and a C-terminal fibronectin type III (FnIII) domain and forms a dimer. The acceptor peptide is sandwiched between the two protomers. The catalytic domain of one protomer recognizes the O-mannosylation site (TPT motif), and the FnIII domain of the other protomer recognizes the C-terminal region of the peptide. Structure-based mutational studies confirmed that amino acid residues of the catalytic domain interacting with mannose or the TPT motif are essential for POMGNT2 enzymatic activity. In addition, the FnIII domain is also essential for the activity and it interacts with the peptide mainly by hydrophobic interaction. Our study provides the first atomic-resolution insights into specific acceptor recognition by the FnIII domain of POMGNT2. The catalytic mechanism of POMGNT2 is proposed based on the structure.
- Molecular Glycobiology, Research Team for Mechanism of Aging, Tokyo Metropolitan Geriatric Hospital and Institute of Gerontology, Itabashi-ku, Japan.
Organizational Affiliation: