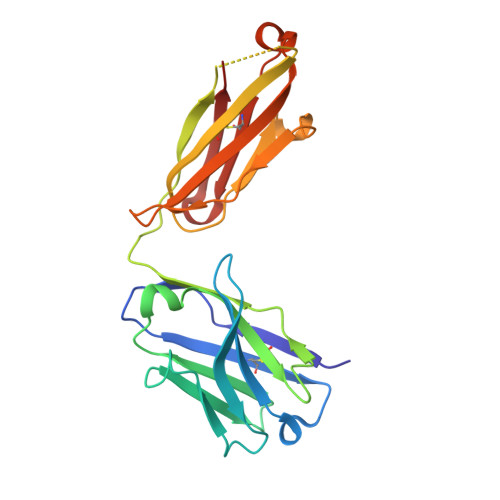
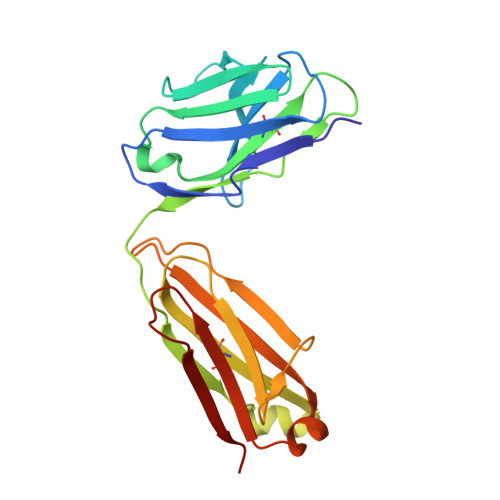
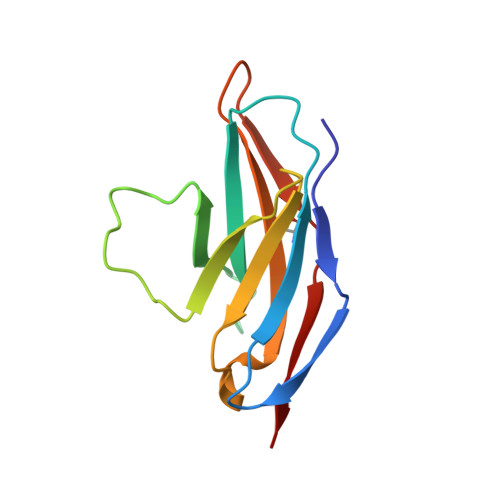
Structural basis of HLX10 PD-1 receptor recognition, a promising anti-PD-1 antibody clinical candidate for cancer immunotherapy.
Issafras, H., Fan, S., Tseng, C.L., Cheng, Y., Lin, P., Xiao, L., Huang, Y.J., Tu, C.H., Hsiao, Y.C., Li, M., Chen, Y.H., Ho, C.H., Li, O., Wang, Y., Chen, S., Ji, Z., Zhang, E., Mao, Y.T., Liu, E., Yang, S., Jiang, W.(2021) PLoS One 16: e0257972-e0257972
- PubMed: 34972111
- DOI: https://doi.org/10.1371/journal.pone.0257972
- Primary Citation of Related Structures:
7E9B - PubMed Abstract:
Cancer immunotherapies, such as checkpoint blockade of programmed cell death protein-1 (PD-1), represents a breakthrough in cancer treatment, resulting in unprecedented results in terms of overall and progression-free survival. Discovery and development of novel anti PD-1 inhibitors remains a field of intense investigation, where novel monoclonal antibodies (mAbs) and novel antibody formats (e.g., novel isotype, bispecific mAb and low-molecular-weight compounds) are major source of future therapeutic candidates. HLX10, a fully humanized IgG4 monoclonal antibody against PD-1 receptor, increased functional activities of human T-cells and showed in vitro, and anti-tumor activity in several tumor models. The combined inhibition of PD-1/PDL-1 and angiogenesis pathways using anti-VEGF antibody may enhance a sustained suppression of cancer-related angiogenesis and tumor elimination. To elucidate HLX10's mode of action, we solved the structure of HLX10 in complex with PD-1 receptor. Detailed epitope analysis showed that HLX10 has a unique mode of recognition compared to the clinically approved PD1 antibodies Pembrolizumab and Nivolumab. Notably, HLX10's epitope was closer to Pembrolizumab's epitope than Nivolumab's epitope. However, HLX10 and Pembrolizumab showed an opposite heavy chain (HC) and light chain (LC) usage, which recognizes several overlapping amino acid residues on PD-1. We compared HLX10 to Nivolumab and Pembrolizumab and it showed similar or better bioactivity in vitro and in vivo, providing a rationale for clinical evaluation in cancer immunotherapy.
- Hengenix Inc., Fremont, CA, United States of America.
Organizational Affiliation: