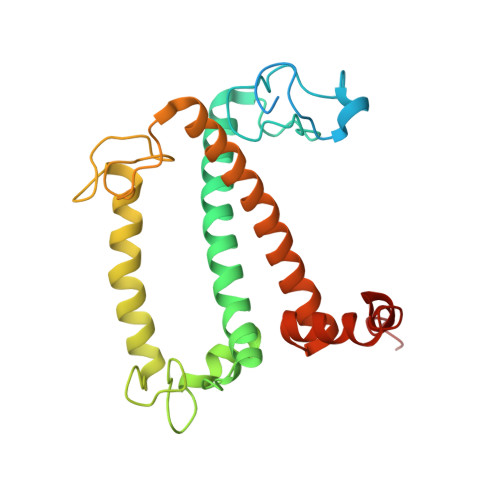
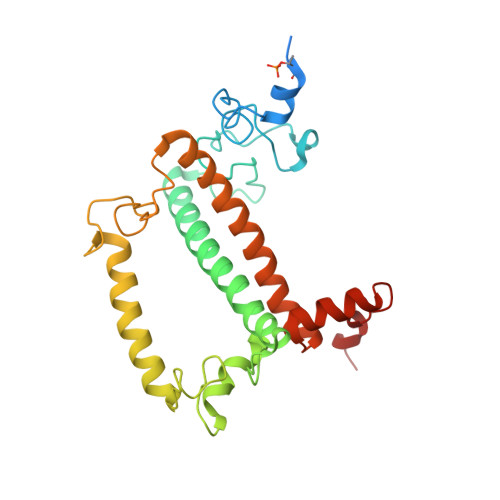
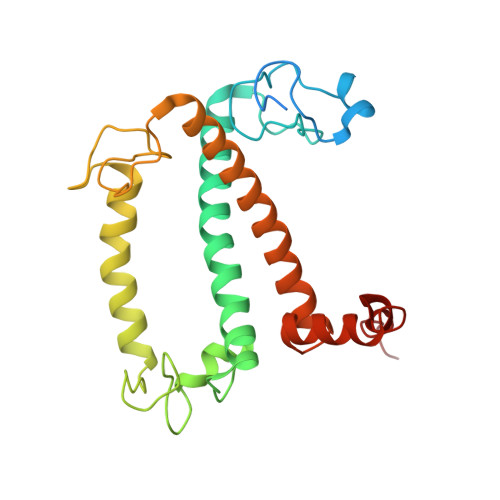
Structural basis of LhcbM5-mediated state transitions in green algae.
Pan, X., Tokutsu, R., Li, A., Takizawa, K., Song, C., Murata, K., Yamasaki, T., Liu, Z., Minagawa, J., Li, M.(2021) Nat Plants 7: 1119-1131
- PubMed: 34239095
- DOI: https://doi.org/10.1038/s41477-021-00960-8
- Primary Citation of Related Structures:
7DZ7, 7DZ8, 7E0H, 7E0I, 7E0J, 7E0K - PubMed Abstract:
In green algae and plants, state transitions serve as a short-term light-acclimation process in the regulation of the light-harvesting capacity of photosystems I and II (PSI and PSII, respectively). During the process, a portion of light-harvesting complex II (LHCII) is phosphorylated, dissociated from PSII and binds with PSI to form the supercomplex PSI-LHCI-LHCII. Here, we report high-resolution structures of PSI-LHCI-LHCII from Chlamydomonas reinhardtii, revealing the mechanism of assembly between the PSI-LHCI complex and two phosphorylated LHCII trimers containing all four types of LhcbM protein. Two specific LhcbM isoforms, namely LhcbM1 and LhcbM5, directly interact with the PSI core through their phosphorylated amino terminal regions. Furthermore, biochemical and functional studies on mutant strains lacking either LhcbM1 or LhcbM5 indicate that only LhcbM5 is indispensable in supercomplex formation. The results unravel the specific interactions and potential excitation energy transfer routes between green algal PSI and two phosphorylated LHCIIs.
- National Laboratory of Biomacromolecules, CAS Centre for Excellence in Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, China.
Organizational Affiliation: