Cross-reactive TCR with alloreactivity for immunodominant HIV-1 epitope Gag TL9 with enhanced control of viral infection
Liu, Y., San, D., Yin, L.(2021) bioRxiv
Experimental Data Snapshot
Starting Models: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MHC class I antigen | 279 | Homo sapiens | Mutation(s): 0 Gene Names: HLA-B |  | |
UniProt | |||||
Find proteins for V6E0U6 (Homo sapiens) Explore V6E0U6 Go to UniProtKB: V6E0U6 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | V6E0U6 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Beta-2-microglobulin | 100 | Homo sapiens | Mutation(s): 0 Gene Names: B2M, CDABP0092, HDCMA22P |  | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P61769 (Homo sapiens) Explore P61769 Go to UniProtKB: P61769 | |||||
PHAROS: P61769 GTEx: ENSG00000166710 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P61769 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar proteins by: Sequence | 3D Structure
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Gag-Pol polyprotein | 9 | Human immunodeficiency virus 1 | Mutation(s): 0 |  | |
UniProt | |||||
Find proteins for P04585 (Human immunodeficiency virus type 1 group M subtype B (isolate HXB2)) Explore P04585 Go to UniProtKB: P04585 | |||||
Entity Groups | |||||
| UniProt Group | P04585 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 4 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| beta chain T18A TCR | 244 | Homo sapiens | Mutation(s): 0 |  | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 5 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| alpha chain T18A TCR | 204 | Homo sapiens | Mutation(s): 0 | 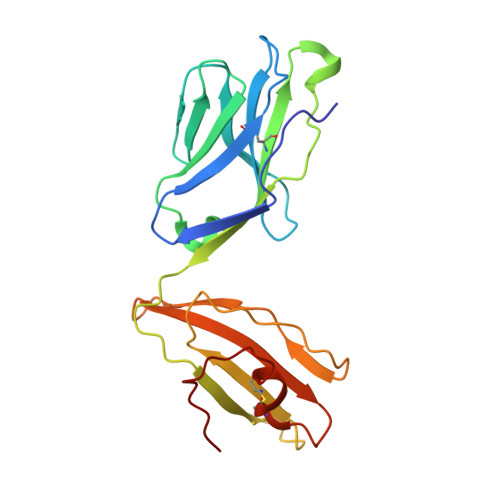 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 96.992 | α = 90 |
| b = 96.992 | β = 90 |
| c = 263.839 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| MOSFLM | data reduction |
| Aimless | data scaling |
| PHASER | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| National Natural Science Foundation of China (NSFC) | China | 31870728 |
| National Natural Science Foundation of China (NSFC) | China | 31470738 |