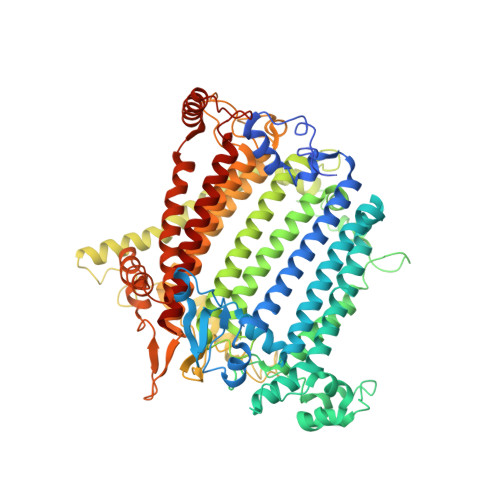
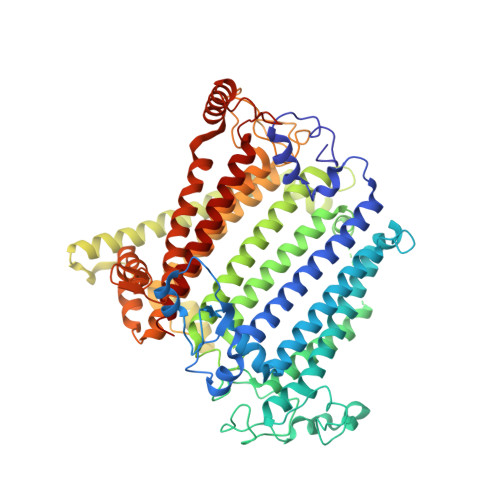
A unique photosystem I reaction center from a chlorophyll d-containing cyanobacterium Acaryochloris marina.
Xu, C., Zhu, Q., Chen, J.H., Shen, L., Yi, X., Huang, Z., Wang, W., Chen, M., Kuang, T., Shen, J.R., Zhang, X., Han, G.(2021) J Integr Plant Biol 63: 1740-1752
- PubMed: 34002536
- DOI: https://doi.org/10.1111/jipb.13113
- Primary Citation of Related Structures:
7DWQ - PubMed Abstract:
Photosystem I (PSI) is a large protein supercomplex that catalyzes the light-dependent oxidation of plastocyanin (or cytochrome c 6 ) and the reduction of ferredoxin. This catalytic reaction is realized by a transmembrane electron transfer chain consisting of primary electron donor (a special chlorophyll (Chl) pair) and electron acceptors A 0 , A 1 , and three Fe 4 S 4 clusters, F X , F A , and F B . Here we report the PSI structure from a Chl d-dominated cyanobacterium Acaryochloris marina at 3.3 Å resolution obtained by single-particle cryo-electron microscopy. The A. marina PSI exists as a trimer with three identical monomers. Surprisingly, the structure reveals a unique composition of electron transfer chain in which the primary electron acceptor A 0 is composed of two pheophytin a rather than Chl a found in any other well-known PSI structures. A novel subunit Psa27 is observed in the A. marina PSI structure. In addition, 77 Chls, 13 α-carotenes, two phylloquinones, three Fe-S clusters, two phosphatidyl glycerols, and one monogalactosyl-diglyceride were identified in each PSI monomer. Our results provide a structural basis for deciphering the mechanism of photosynthesis in a PSI complex with Chl d as the dominating pigments and absorbing far-red light.
- Department of Biophysics and Department of Pathology of Sir Run Run Shaw Hospital, Zhejiang University School of Medicine, Hangzhou, 310058, China.
Organizational Affiliation: