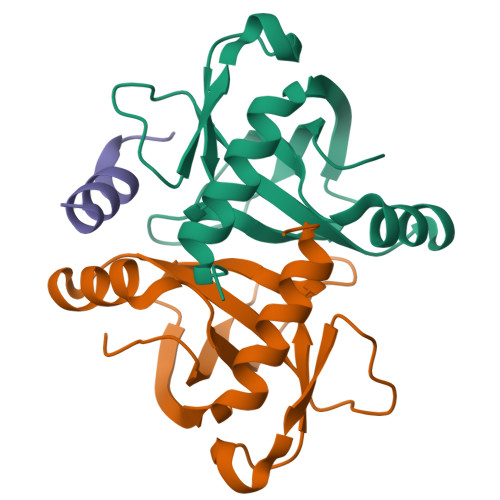
Mechanistic Insight into the Peptide Binding Modes to Two M. tb MazF Toxins.
Chen, R., Zhou, J., Xie, W.(2021) Toxins (Basel) 13
- PubMed: 33925254
- DOI: https://doi.org/10.3390/toxins13050319
- Primary Citation of Related Structures:
7DU4, 7DU5 - PubMed Abstract:
Tuberculosis (TB) is a contagious disease caused by Mycobacterium tuberculosis ( M. tb ). It is regarded as a major health threat all over the world, mainly because of its high mortality and drug-resistant nature. Toxin-antitoxin (TA) systems are modules ubiquitously found in prokaryotic organisms, and the well-studied MazEF systems (MazE means "what is it?" in Hebrew) are implicated in the formation of "persister cells" in the M. tb pathogen. Here, we report cocrystal structures of M. tb MazF-mt1 and -mt9, two important MazF members responsible for specific mRNA and tRNA cleavages, respectively, in complexes with truncated forms of their cognate antitoxin peptides. These peptides bind to the toxins with comparable affinities to their full-length antitoxins, which would reduce the RNA-cleavage capacities of the toxins in vitro. After structural analysis of the binding modes, we systemically tested the influence of the substitutions of individual residues in the truncated MazE-mt9 peptide on its affinity. This study provides structural insight into the binding modes and the inhibition mechanisms between the MazE/F-mt TA pairs. More importantly, it contributes to the future design of peptide-based antimicrobial agents against TB and potentially relieves the drug-resistance problems by targeting novel M. tb proteins.
Organizational Affiliation:
MOE Key Laboratory of Gene Function and Regulation, State Key Laboratory for Biocontrol, School of Life Sciences, The Sun Yat-Sen University, Guangzhou 510006, China.