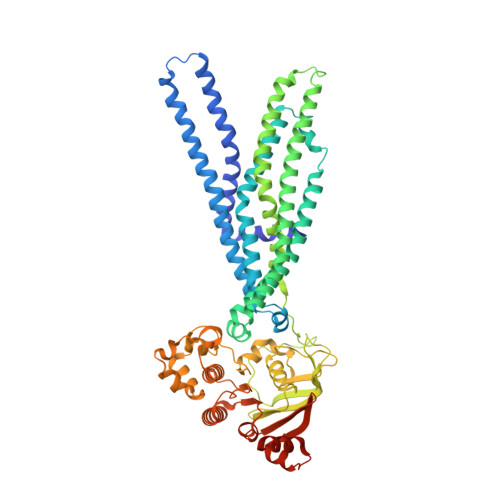
The crystal structure of the CmABCB1 G132V mutant, which favors the outward-facing state, reveals the mechanism of the pivotal joint between TM1 and TM3.
Matsuoka, K., Nakatsu, T., Kato, H.(2021) Protein Sci 30: 1064-1071
- PubMed: 33683740
- DOI: https://doi.org/10.1002/pro.4058
- Primary Citation of Related Structures:
7DQV - PubMed Abstract:
CmABCB1 is a homologue of human P-glycoprotein, which extrudes various substrates by iterative cycles of conformational changes between the inward- and outward-facing states. Comparison of the inward- and outward-facing structures of CmABCB1 suggested that pivotal joints in the transmembrane domain regulate the tilt of transmembrane helices. Transmembrane helix 1 (TM1) forms a tight helix-helix contact with TM3 at the TM1-3 joint. Mutation of Gly132 to valine at the TM1-3 joint, G132V, caused a 10-fold increase in ATPase activity, but the mechanism underlying this change remains unclear. Here, we report a crystal structure of the outward-facing state of the CmABCB1 G132V mutant at a 2.15 Å resolution. We observed structural displacements between the outward-facing states of G132V and the previous one at the region around the TM1-3 joint, and a significant expansion at the extracellular gate. We hypothesize that steric hindrance caused by the Val substitution shifted the conformational equilibrium toward the outward-facing state, favoring the dimeric state of the nucleotide-binding domains and thereby increasing the ATPase activity of the G132V mutant.
- Department of Structural Biology, Graduate School of Pharmaceutical Sciences, Kyoto University, Kyoto, Japan.
Organizational Affiliation: