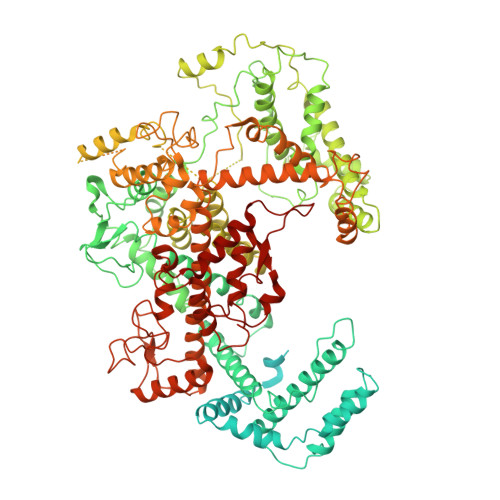
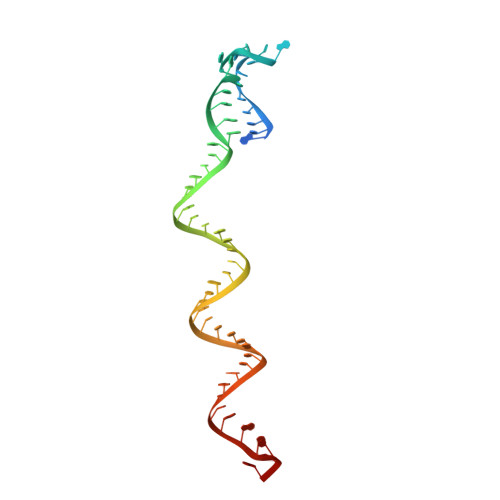
Structural basis for self-cleavage prevention by tag:anti-tag pairing complementarity in type VI Cas13 CRISPR systems.
Wang, B., Zhang, T., Yin, J., Yu, Y., Xu, W., Ding, J., Patel, D.J., Yang, H.(2021) Mol Cell 81: 1100
- PubMed: 33472057
- DOI: https://doi.org/10.1016/j.molcel.2020.12.033
- Primary Citation of Related Structures:
7DMQ - PubMed Abstract:
Bacteria and archaea apply CRISPR-Cas surveillance complexes to defend against foreign invaders. These invading genetic elements are captured and integrated into the CRISPR array as spacer elements, guiding sequence-specific DNA/RNA targeting and cleavage. Recently, in vivo studies have shown that target RNAs with extended complementarity with repeat sequences flanking the target element (tag:anti-tag pairing) can dramatically reduce RNA cleavage by the type VI-A Cas13a system. Here, we report the cryo-EM structure of Leptotrichia shahii LshCas13a crRNA in complex with target RNA harboring tag:anti-tag pairing complementarity, with the observed conformational changes providing a molecular explanation for inactivation of the composite HEPN domain cleavage activity. These structural insights, together with in vitro biochemical and in vivo cell-based assays on key mutants, define the molecular principles underlying Cas13a's capacity to target and discriminate between self and non-self RNA targets. Our studies illuminate approaches to regulate Cas13a's cleavage activity, thereby influencing Cas13a-mediated biotechnological applications.
- State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, Chinese Academy of Sciences, University of Chinese Academy of Sciences, Shanghai 200031, China.
Organizational Affiliation: