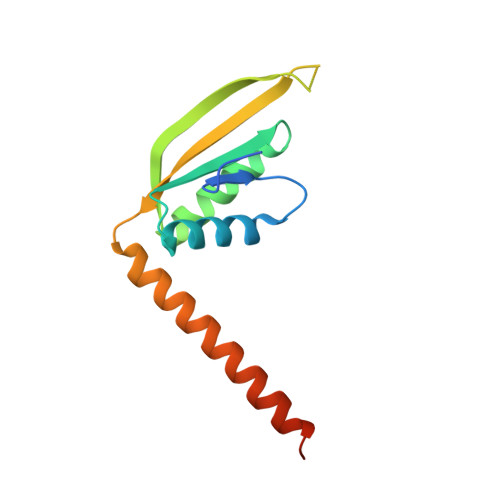
Crystal structure of TbAlba1 from Trypanosoma brucei.
Gao, J., Xiao, C., Liao, S., Tu, X.(2021) J Struct Biol 213: 107751-107751
- PubMed: 34107324
- DOI: https://doi.org/10.1016/j.jsb.2021.107751
- Primary Citation of Related Structures:
7DL8 - PubMed Abstract:
Alba (Acetylation lowers binding affinity) domain is a small, dimeric nucleic acid-binding domain, which is widely distributed in archaea and numbers of eukaryotes. Alba domain containing proteins have been reported to be involved in many cellular processes, such as regulation of translation, maintaining genome stability, regulation of RNA processing and so on. In Trypanosoma brucei (T. brucei), there are four Alba proteins identified, which are named TbAlba1 to TbAlba4. However, the structure and function of TbAlba proteins are still unknown. Here, we solved the crystal structure of TbAlba1 to a resolution of 2.46 Å. TbAlba1 adopts a similar Alba-fold, which comprises of four β-strands (β1-β4) and three long α-helices (α1-α3). Furthermore, TbAlba1 displays some structural features quite different from other Alba proteins. These differences may imply the diverse biological roles of Alba family members.
- Hefei National Laboratory for Physical Sciences at Microscale, and School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, PR China; Department of Ophthalmology, The Second Affiliated Hospital of Anhui Medical University, 678 Furong Road, Hefei, Anhui, PR China.
Organizational Affiliation: