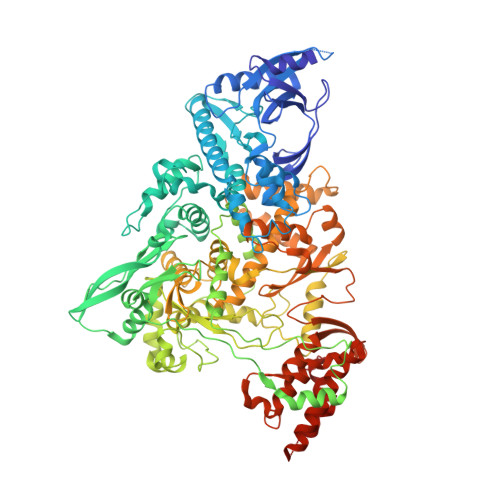
Structural basis for repurpose and design of nucleotide drugs for treating COVID-19
Yin, W., Luan, X., Li, Z., Xie, Y., Zhou, Z., Liu, J., Gao, M., Wang, X., Zhou, F., Wang, Q., Wang, Q., Shen, D., Zhang, Y., Tian, G., Aisa, H., Wei, D., Jiang, Y., Xiao, G., Jiang, H., Zhang, L., Yu, X., Shen, J., Zhang, S., Xu, H.To be published.