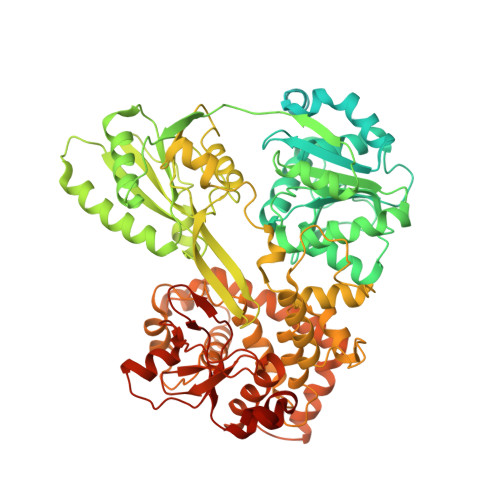
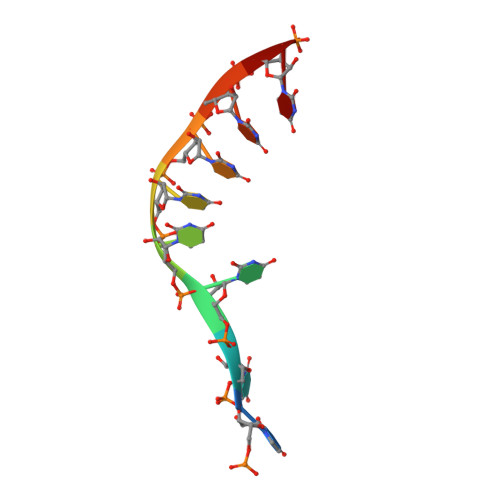
Mechanism of spliceosome remodeling by the ATPase/helicase Prp2 and its coactivator Spp2.
Bai, R., Wan, R., Yan, C., Jia, Q., Lei, J., Shi, Y.(2021) Science 371
- PubMed: 33243853
- DOI: https://doi.org/10.1126/science.abe8863
- Primary Citation of Related Structures:
7DCO, 7DCP, 7DCQ, 7DCR, 7DD3 - PubMed Abstract:
Spliceosome remodeling, executed by conserved adenosine triphosphatase (ATPase)/helicases including Prp2, enables precursor messenger RNA (pre-mRNA) splicing. However, the structural basis for the function of the ATPase/helicases remains poorly understood. Here, we report atomic structures of Prp2 in isolation, Prp2 complexed with its coactivator Spp2, and Prp2-loaded activated spliceosome and the results of structure-guided biochemical analysis. Prp2 weakly associates with the spliceosome and cannot function without Spp2, which stably associates with Prp2 and anchors on the spliceosome, thus tethering Prp2 to the activated spliceosome and allowing Prp2 to function. Pre-mRNA is loaded into a featured channel between the N and C halves of Prp2, where Leu 536 from the N half and Arg 844 from the C half prevent backward sliding of pre-mRNA toward its 5'-end. Adenosine 5'-triphosphate binding and hydrolysis trigger interdomain movement in Prp2, which drives unidirectional stepwise translocation of pre-mRNA toward its 3'-end. These conserved mechanisms explain the coupling of spliceosome remodeling to pre-mRNA splicing.
- Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Hangzhou 310024, Zhejiang Province, China.
Organizational Affiliation: