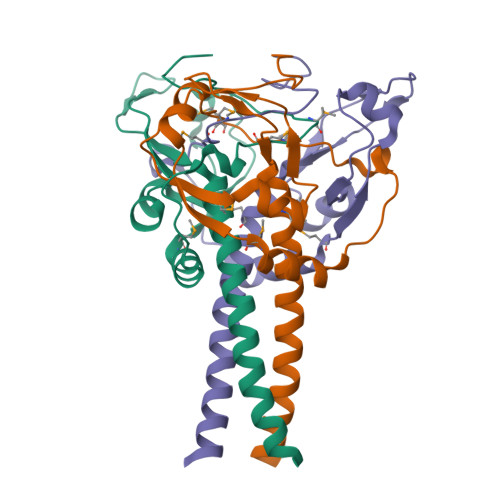
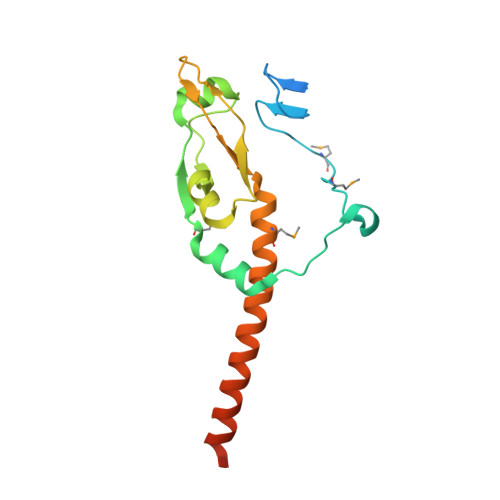
Crystal structure of the MyRF ICA domain with its upstream beta-helical stalk reveals the molecular mechanisms underlying its trimerization and self-cleavage.
Wu, P., Zhen, X., Li, B., Yu, Q., Huang, X., Shi, N.(2021) Int J Biol Sci 17: 2931-2943
- PubMed: 34345217
- DOI: https://doi.org/10.7150/ijbs.57673
- Primary Citation of Related Structures:
7DC3 - PubMed Abstract:
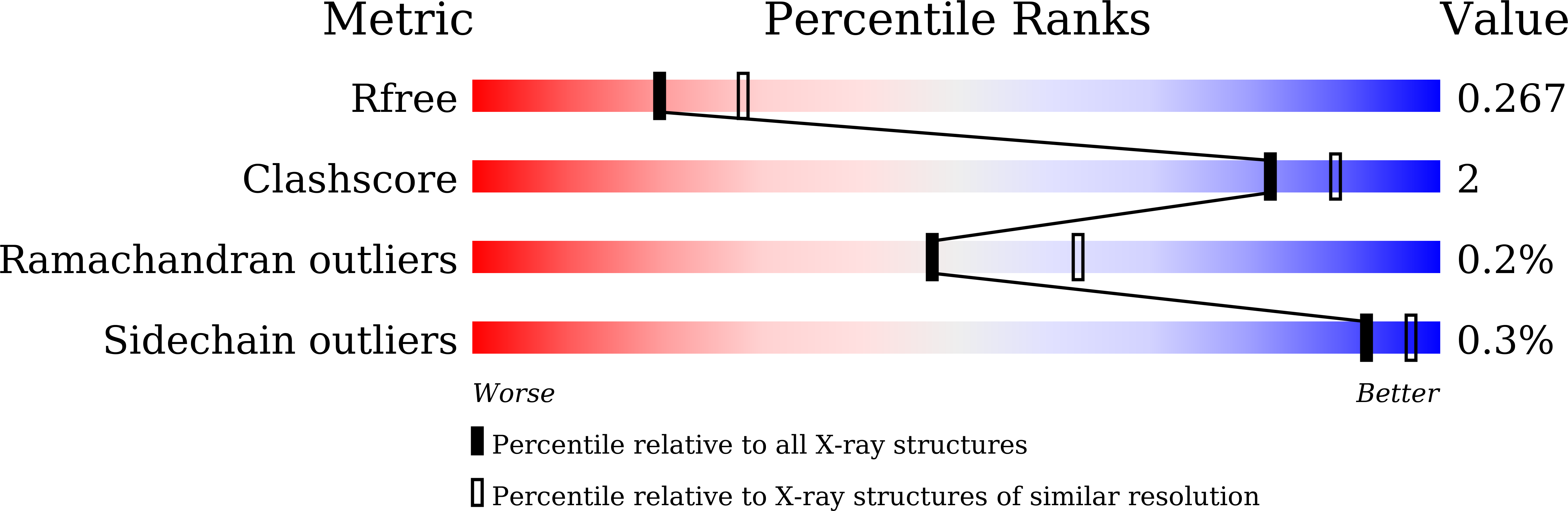
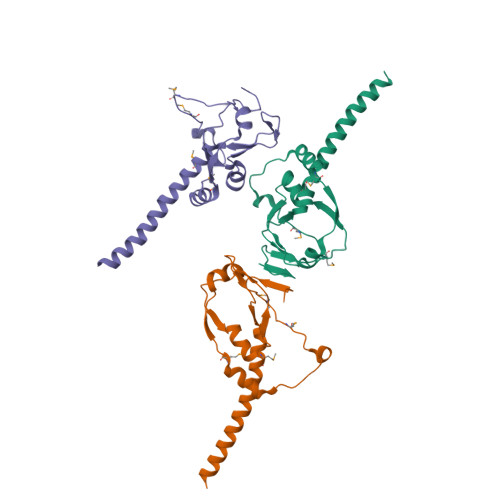
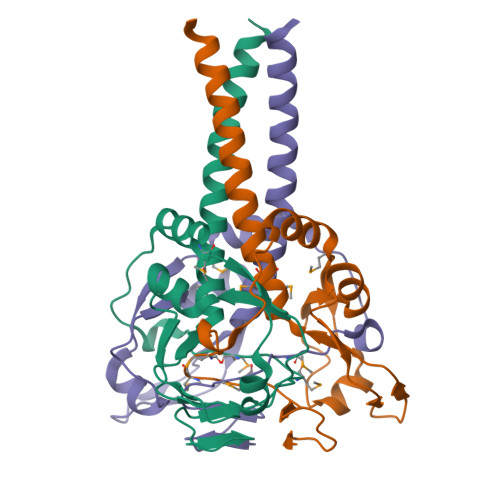
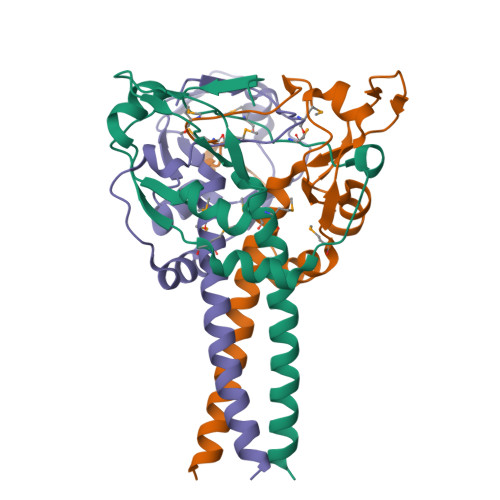
Myelin gene regulatory factor (MyRF), a novel membrane transcription factor expressed on the endoplasmic reticulum membrane, functions as a trimer. The trimerization of MyRF is associated with a fragment between the DNA binding domain and transmembrane domain that shares homology with the triple-β-helix and intramolecular chaperone autocleavage (ICA) domain of phage tailspike proteins. The molecular details of these domains in eukaryotes have not been elucidated. Here, we present the crystal structure of the MyRF ICA domain with its upstream β-helical stalk, determined at 2.4Å resolution. The structure showed that its upstream β-helical stalk is different from the triple β-helix reported before. This is the first structure of the mammalian protein with a triple β-helix. Structure analysis demonstrated that the triple α-helical coiled-coil formed at the MyRF ICA domain C-terminal was the main driving force for the trimerization. Additionally, our findings showed that MyRF was cleaved via a highly conserved serine-lysine catalytic dyad mechanism and that cleavage would be activated only if the ICA domains were organized as trimers. In contrast to the viral ICA domain, almost no interaction was found between the MyRF ICA domain and its upstream neighboring β-helix of the stalk; thus, activation of self-cleavage may not be triggered by the upstream region of the ICA domain, contrary to the observations made in phages. These findings provided an important insight into the molecular mechanisms of MyRF trimerization and self-cleavage.
Organizational Affiliation:
State Key Laboratory of Structural Chemistry, Fujian Institute of Research on the Structure of Matter, Chinese Academy of Sciences, Fujian College, University of Chinese Academy of Sciences, 155 Yangqiao Road West, Fuzhou, 350002, China.