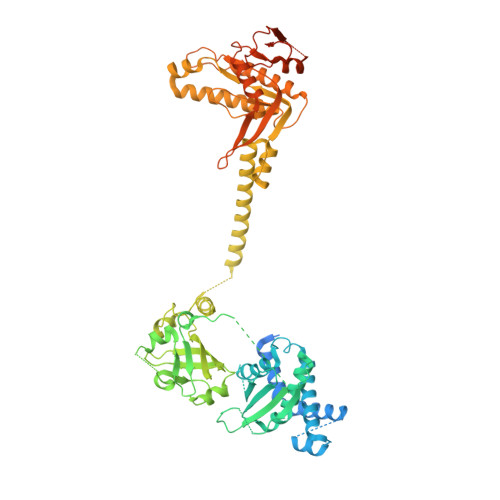
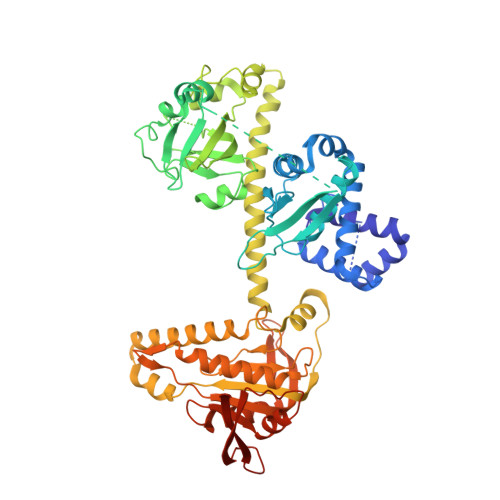
Activation mechanism of human soluble guanylate cyclase by stimulators and activators.
Liu, R., Kang, Y., Chen, L.(2021) Nat Commun 12: 5492-5492
- PubMed: 34535643
- DOI: https://doi.org/10.1038/s41467-021-25617-0
- Primary Citation of Related Structures:
7D9R, 7D9S, 7D9T, 7D9U - PubMed Abstract:
Soluble guanylate cyclase (sGC) is the receptor for nitric oxide (NO) in human. It is an important validated drug target for cardiovascular diseases. sGC can be pharmacologically activated by stimulators and activators. However, the detailed structural mechanisms, through which sGC is recognized and positively modulated by these drugs at high spacial resolution, are poorly understood. Here, we present cryo-electron microscopy structures of human sGC in complex with NO and sGC stimulators, YC-1 and riociguat, and also in complex with the activator cinaciguat. These structures uncover the molecular details of how stimulators interact with residues from both β H-NOX and CC domains, to stabilize sGC in the extended active conformation. In contrast, cinaciguat occupies the haem pocket in the β H-NOX domain and sGC shows both inactive and active conformations. These structures suggest a converged mechanism of sGC activation by pharmacological compounds.
- State Key Laboratory of Membrane Biology, College of Future Technology, Institute of Molecular Medicine, Beijing Key Laboratory of Cardiometabolic Molecular Medicine, Peking University, 100871, Beijing, China.
Organizational Affiliation: