Kinetics-Driven Drug Design Strategy for Next-Generation Acetylcholinesterase Inhibitors to Clinical Candidate.
Zhou, Y., Fu, Y., Yin, W., Li, J., Wang, W., Bai, F., Xu, S., Gong, Q., Peng, T., Hong, Y., Zhang, D., Zhang, D., Liu, Q., Xu, Y., Xu, H.E., Zhang, H., Jiang, H., Liu, H.(2021) J Med Chem 64: 1844-1855
- PubMed: 33570950
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01863
- Primary Citation of Related Structures:
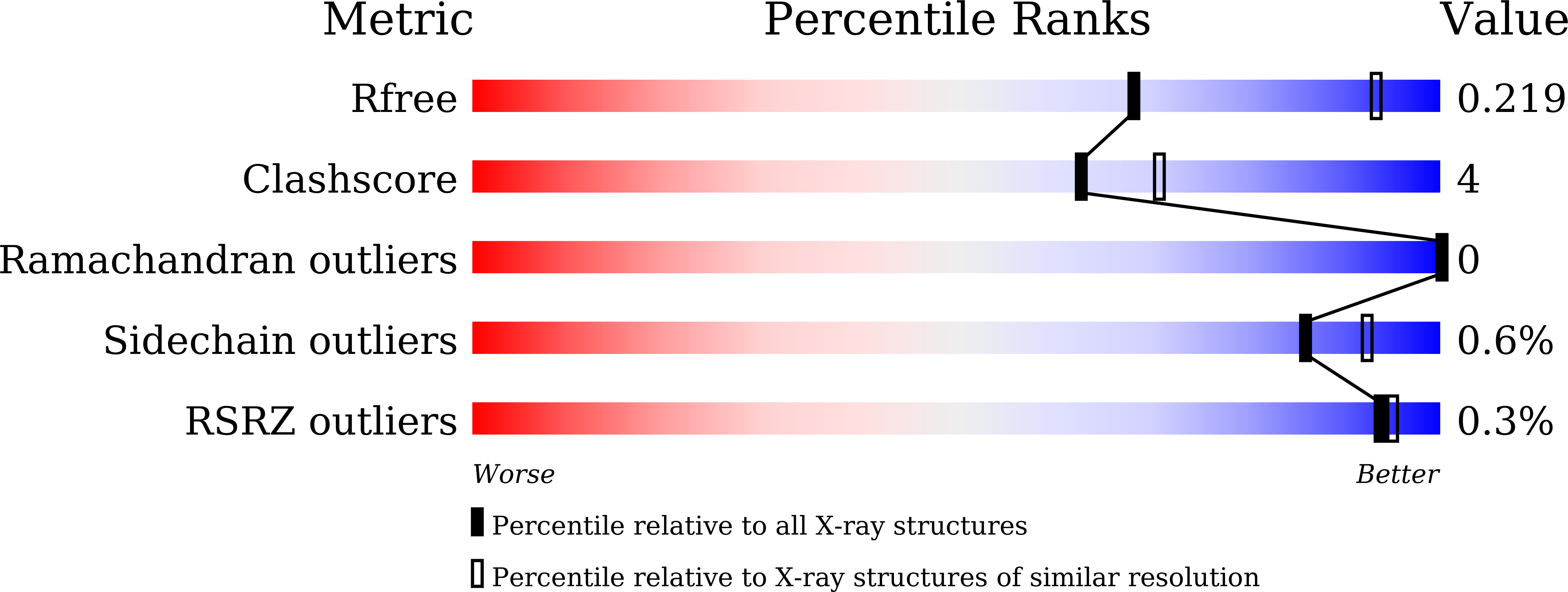
7D9O, 7D9P, 7D9Q - PubMed Abstract:
The acetylcholinesterase (AChE) inhibitors remain key therapeutic drugs for the treatment of Alzheimer's disease (AD). However, the low-safety window limits their maximum therapeutic benefits. Here, a novel kinetics-driven drug design strategy was employed to discover new-generation AChE inhibitors that possess a longer drug-target residence time and exhibit a larger safety window. After detailed investigations, compound 12 was identified as a highly potent, highly selective, orally bioavailable, and brain preferentially distributed AChE inhibitor. Moreover, it significantly ameliorated cognitive impairments in different mouse models with a lower effective dose than donepezil. The X-ray structure of the cocrystal complex provided a precise binding mode between 12 and AChE. Besides, the data from the phase I trials demonstrated that 12 had good safety, tolerance, and pharmacokinetic profiles at all preset doses in healthy volunteers, providing a solid basis for its further investigation in phase II trials for the treatment of AD.
Organizational Affiliation:
State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zu Chong Zhi Road, Shanghai 201203, People's Republic of China.